Hi, I am working with one set of anatomical and functional run from my lab. I am using Heudiconv and after converting my dataset, I realized that I am not getting theINFO:[node]finished “embedder” message.
INFO: [Node] Finished “convert”.
200114-12:53:36,536 nipype.workflow INFO:
[Node] Setting-up “embedder” in “/tmp/embedmetaher4eo8k/embedder”.
INFO: [Node] Setting-up “embedder” in “/tmp/embedmetaher4eo8k/embedder”.
200114-12:54:39,405 nipype.workflow INFO:
[Node] Running “embedder” (“nipype.interfaces.utility.wrappers.Function”)
INFO: [Node] Running “embedder” (“nipype.interfaces.utility.wrappers.Function”)
(base) ndsl-eeg-9513:~ abdollahis2$
however, looking at the file structure, I get the BIDS formatted dataset.so I move on to the next step to validate my dataset and I get the following error:
I have read that one way to fix this issue is to manually add TaskName type (i.e rest) to the json file, but I was wondering if there is a better way to fix it?
Thanks
Shervin
Hi @shervin.abd71
Thank you for your message! It appears the dataset looks good other than what you described (adding TaskName). We put together a tutorial and on step 11 we have shown how to insert the TaskName programmatically using jq. The lines of interest happen in the code block under #Does TaskName exist?
Thank you,
Franklin
Thanks Franklin for your suggestion, however I am following the website’s nipy/HeudiConv pipeline and for that I have a heuristic.py scripts and a simple command line script (where I run the docker commands). can I add that piece of code to either scripts and have the TaskName added to son file?
Hi @shervin.abd71
It appears this can be achieved within HeuDiConv. We have another tutorial focused on using HeuDiConv that may be useful to look at! It looks like it is within the heuristic.py script with no additional scripting needed
Thank you,
Franklin
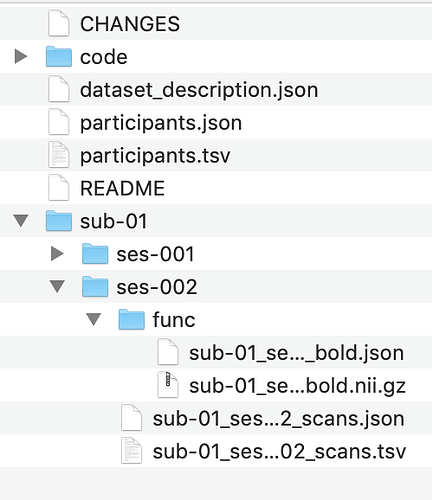
I am following that same page and if you look at my code its exactly the same as the tutorial. however, I am getting wrongly name json file on my functional runs (ses-002). I am not sure where I am going wrong?
Hi @shervin.abd71
Thank you for sharing your code! It appears there is a top-level task-rest_bold.json (this pops up at step 7) that is causing this error. May you please share the command you are using to generate these files?
Thank you,
Franklin
Sure, here are the commands I am using.
Also, step 7 mentions, deleting dicominfo.tsv (assuming its dicominfo_ses-002.tsv) file (that I have copied to my base directory) before running the docker command for dcm2niix convertor. for this run I haven’t removed the tsv file from my base directory, do you think that where the problem arises?
that could be it! I recall when putting it together being careful of the hidden .heudiconv folder as well (at least when this put together).
the commands look good
its not that. I tried it again, i got the same error. However, I recently found out that I am dealing with multicho fMRI dataset, where for my Raw_Dicom_folder, I have 3 different README.txt, README-Series_v002.txt, and README-Series_v003.txt files. Is Heudiconv capable to identifying the difference?
There appears to be some work extending HeuDiConv into multiecho - this could be a reason why the top level task json file was not being generated. cc: @mgxd if he may have some tips!
Thanks franklin, i have read the available discussions on multiecho fmri scans by mgxd and will get in touch with him regarding to my problem.