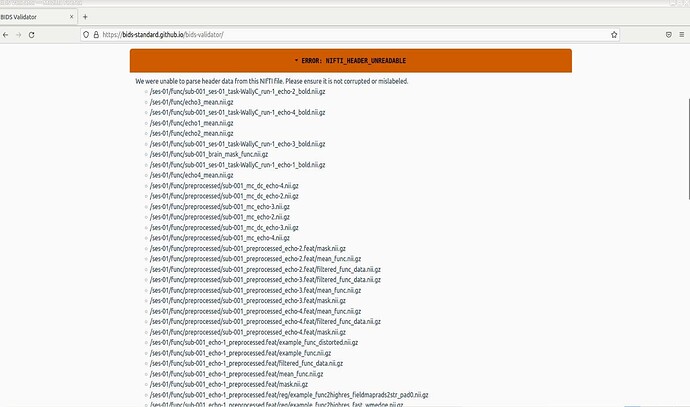
Hi,
I’ve converted my DICOMs to BIDS and tried to run it through the online BIDs validator. I’m receiving this error: NIFTI_HEADER_UNREADABLE, for quite a few of my files. Here is fslhd performed on one of the identified files:
filename sub-001_ses-01_task-WallyC_run-1_echo-1_bold.nii.gz
sizeof_hdr 348
data_type UINT16
dim0 4
dim1 64
dim2 64
dim3 48
dim4 653
dim5 1
dim6 1
dim7 1
vox_units mm
time_units s
datatype 512
nbyper 2
bitpix 16
pixdim0 -1.000000
pixdim1 3.000000
pixdim2 3.000000
pixdim3 3.000000
pixdim4 2.000000
pixdim5 0.000000
pixdim6 0.000000
pixdim7 0.000000
vox_offset 352
cal_max 0.000000
cal_min 0.000000
scl_slope 1.000000
scl_inter 0.000000
phase_dim 2
freq_dim 1
slice_dim 3
slice_name Unknown
slice_code 0
slice_start 0
slice_end 0
slice_duration 0.000000
toffset 0.000000
intent Unknown
intent_code 0
intent_name
intent_p1 0.000000
intent_p2 0.000000
intent_p3 0.000000
qform_name Scanner Anat
qform_code 1
qto_xyz:1 -2.995243 -0.031890 0.165836 98.091675
qto_xyz:2 -0.056566 2.965300 -0.451441 -76.222855
qto_xyz:3 0.159119 0.453852 2.961199 -67.908783
qto_xyz:4 0.000000 0.000000 0.000000 1.000000
qform_xorient Right-to-Left
qform_yorient Posterior-to-Anterior
qform_zorient Inferior-to-Superior
sform_name Scanner Anat
sform_code 1
sto_xyz:1 -2.995243 -0.031890 0.165834 98.091675
sto_xyz:2 -0.056566 2.965299 -0.451441 -76.222855
sto_xyz:3 0.159117 0.453852 2.961199 -67.908783
sto_xyz:4 0.000000 0.000000 0.000000 1.000000
sform_xorient Right-to-Left
sform_yorient Posterior-to-Anterior
sform_zorient Inferior-to-Superior
file_type NIFTI-1+
file_code 1
descrip TE=12;Time=150256.575;phase=1;mb=2
aux_file Unaliased MB2/PE4/LB
Just for some context, this is a multi-echo acquisition. I used dcm2bids version: 3.2.0, based on BIDS version: v1.9.0.
Any insights would be greatly appreciated!