Hi,
I have used heudiconv to convert a DICOM dataset into NIfTI files/BIDS layout.
So the layout is the following:
Investigators/MyStudy
Investigators/MyStudyFDNeuro08242021
Investigators/MyStudyFDPain08242021
Investigators/MyStudySZPain
Investigators/MyStudySZPain10820
Investigators/MyStudySZPain8232021
each having its own BIDS structure files (CHANGES , dataset_description.json , participants.json , participants.tsv , README , etc.).
I am trying to validate the data with BIDS-Validator installed through npm (version 1.14.13).
Although each study has a .bidsignore file I had to add a .bidsignore file to Investigators to skip the **/sourcedata and **/.heudiconv folders within each study and sourcedata folder within each study.
I have tried both the command line and the browser version of the BIDS-Validator:
- When I call
bids-validator /mnt/data/Investigatorsfrom the command line I get a number of errors that I assume I should interpret as telling that I cannot run the validator on the root folder:
$ bids-validator /mnt/data/Investigators
bids-validator@1.14.13
(node:2783439) Warning: Closing directory handle on garbage collection
(Use `node --trace-warnings ...` to show where the warning was created)
1: [ERR] Files with such naming scheme are not part of BIDS
specification. This error is most commonly caused by typos in file names
that make them not BIDS compatible. Please consult the specification and
make sure your files are named correctly. If this is not a file naming issue
(for example when including files not yet covered by the BIDS specification)
you should include a ".bidsignore" file in your dataset (see
https://github.com/bids-standard/bids-validator#bidsignore for details).
Please note that derived (processed) data should be placed in /derivatives
folder and source data (such as DICOMS or behavioural logs in proprietary
formats) should be placed in the /sourcedata folder. (code:
1 - NOT_INCLUDED)
./MyStudy/CHANGES
Evidence: CHANGES
./MyStudy/README
Evidence: README
./MyStudy/dataset_description.json
Evidence: dataset_description.json
./MyStudy/participants.json
Evidence: participants.json
./MyStudy/participants.tsv
Evidence: participants.tsv
./MyStudy/scans.json
Evidence: scans.json
./MyStudy/sub-263189/anat/sub-263189_echo-1_T1w.json
Evidence: sub-263189_echo-1_T1w.json
./MyStudy/sub-263189/anat/sub-263189_echo-1_T1w.nii.gz
Evidence: sub-263189_echo-1_T1w.nii.gz
./MyStudy/sub-263189/anat/sub-263189_echo-2_T1w.json
Evidence: sub-263189_echo-2_T1w.json
./MyStudy/sub-263189/anat/sub-263189_echo-2_T1w.nii.gz
Evidence: sub-263189_echo-2_T1w.nii.gz
... and 1433 more files having this issue (Use --verbose to see them all).
2: [ERR] There are no subject folders (labeled "sub-*") in the root of this dataset. (code: 45 - SUBJECT_FOLDERS)
3: [ERR] The compulsory file /dataset_description.json is missing. See Section 03 (Modality agnostic files) of the BIDS specification. (code: 57 - DATASET_DESCRIPTION_JSON_MISSING)
4: [ERR] Sub label contain an Illegal Character hyphen or underscore. Please edit the filename as per BIDS spec. (code: 62 - SUBJECT_VALUE_CONTAINS_ILLEGAL_CHARACTER)
./MyStudySZPain10820/sub-155728/anat/sub-155728_T1w_heudiconv323_e1_Eq_1.nii.gz
Evidence: sub name contains illegal character:/MyStudySZPain10820/sub-155728/anat/sub-155728_T1w_heudiconv323_e1_Eq_1.nii.gz
./MyStudySZPain10820/sub-155728/anat/sub-155728_T1w_heudiconv323_e3_Eq_1.nii.gz
5: [ERR] Subject label in the filename doesn't match with the path of the file. File seems to be saved in incorrect subject directory. (code: 64 - SUBJECT_LABEL_IN_FILENAME_DOESNOT_MATCH_DIRECTORY)
./MyStudy/sub-263189/anat/sub-263189_echo-1_T1w.json
Evidence: File: /MyStudy/sub-263189/anat/sub-263189_echo-1_T1w.json is saved in incorrect subject directory as per sub-id in filename.
./MyStudy/sub-263189/anat/sub-263189_echo-1_T1w.nii.gz
Evidence: File: /MyStudy/sub-263189/anat/sub-263189_echo-1_T1w.nii.gz is saved in incorrect subject directory as per sub-id in filename.
./MyStudy/sub-263189/anat/sub-263189_echo-2_T1w.json
Evidence: File: /MyStudy/sub-263189/anat/sub-263189_echo-2_T1w.json is saved in incorrect subject directory as per sub-id in filename.
./MyStudy/sub-263189/anat/sub-263189_echo-2_T1w.nii.gz
Evidence: File: /MyStudy/sub-263189/anat/sub-263189_echo-2_T1w.nii.gz is saved in incorrect subject directory as per sub-id in filename.
./MyStudy/sub-263189/anat/sub-263189_echo-3_T1w.json
Evidence: File: /MyStudy/sub-263189/anat/sub-263189_echo-3_T1w.json is saved in incorrect subject directory as per sub-id in filename.
./MyStudy/sub-263189/anat/sub-263189_echo-3_T1w.nii.gz
Evidence: File: /MyStudy/sub-263189/anat/sub-263189_echo-3_T1w.nii.gz is saved in incorrect subject directory as per sub-id in filename.
./MyStudy/sub-263189/anat/sub-263189_echo-4_T1w.json
Evidence: File: /MyStudy/sub-263189/anat/sub-263189_echo-4_T1w.json is saved in incorrect subject directory as per sub-id in filename.
./MyStudy/sub-263189/anat/sub-263189_echo-4_T1w.nii.gz
Evidence: File: /MyStudy/sub-263189/anat/sub-263189_echo-4_T1w.nii.gz is saved in incorrect subject directory as per sub-id in filename.
./MyStudy/sub-263189/dwi/sub-263189_acq-dir99_dir-ap_dwi.bval
Evidence: File: /MyStudy/sub-263189/dwi/sub-263189_acq-dir99_dir-ap_dwi.bval is saved in incorrect subject directory as per sub-id in filename.
./MyStudy/sub-263189/dwi/sub-263189_acq-dir99_dir-ap_dwi.bvec
Evidence: File: /MyStudy/sub-263189/dwi/sub-263189_acq-dir99_dir-ap_dwi.bvec is saved in incorrect subject directory as per sub-id in filename.
... and 1397 more files having this issue (Use --verbose to see them all).
6: [ERR] No BIDS compatible data found for at least one subject. (code: 67 - NO_VALID_DATA_FOUND_FOR_SUBJECT)
./sub-092743
./sub-139240
(...)
... and 57 more files having this issue (Use --verbose to see them all).
1: [WARN] The recommended file /README is missing. See Section 03 (Modality agnostic files) of the BIDS specification. (code: 101 - README_FILE_MISSING)
Summary: Available Tasks: Available Modalities:
1443 Files, 34.03GB
0 - Subjects
1 - Session
- If I run the command line instruction on each study e.g.
bids-validator /mnt/data/Investigators/MyStudy, I get an error that I cannot understand/debug:
$ bids-validator /mnt/data/Investigators/MyStudy
bids-validator@1.14.13
Unhandled rejection (
reason: Error: strict mode: unknown keyword: "dependency-definitions"
at checkStrictMode (/usr/lib/node_modules/bids-validator/dist/commonjs/cli.js:89219:15)
at checkUnknownRules (/usr/lib/node_modules/bids-validator/dist/commonjs/cli.js:89092:11)
at checkKeywords (/usr/lib/node_modules/bids-validator/dist/commonjs/cli.js:90443:36)
at validateFunctionCode (/usr/lib/node_modules/bids-validator/dist/commonjs/cli.js:90346:9)
at Ajv2.compileSchema (/usr/lib/node_modules/bids-validator/dist/commonjs/cli.js:90944:45)
at Ajv2._compileSchemaEnv (/usr/lib/node_modules/bids-validator/dist/commonjs/cli.js:92601:35)
at Ajv2.compile (/usr/lib/node_modules/bids-validator/dist/commonjs/cli.js:92309:37)
at validateSchema (/usr/lib/node_modules/bids-validator/dist/commonjs/cli.js:104315:28)
at checkUnits (/usr/lib/node_modules/bids-validator/dist/commonjs/cli.js:104231:28)
at json_default2 (/usr/lib/node_modules/bids-validator/dist/commonjs/cli.js:104223:30)
).
I assume that given that each subfolder is a BIDS dataset, the validator should do its job and analyze the data, so I do not know why I am getting the above error.
- If I use the browser to validate the dataset, giving
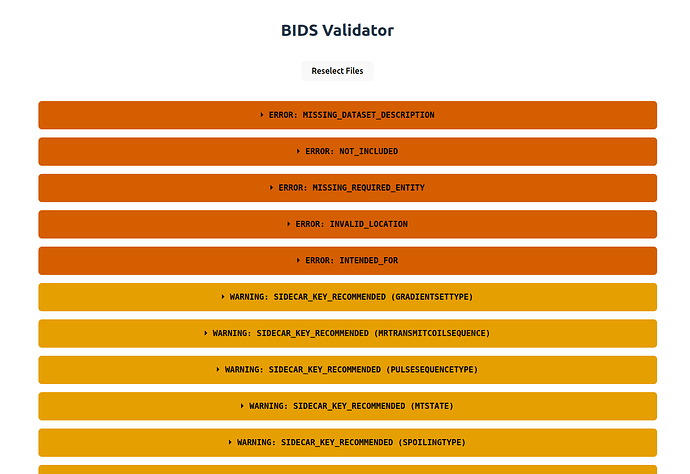
/mnt/data/Investigatorsresults in a number of errors (besides the warnings) that I assume mean that this is not a BIDS dataset (missingdataset_description.json, etc.):
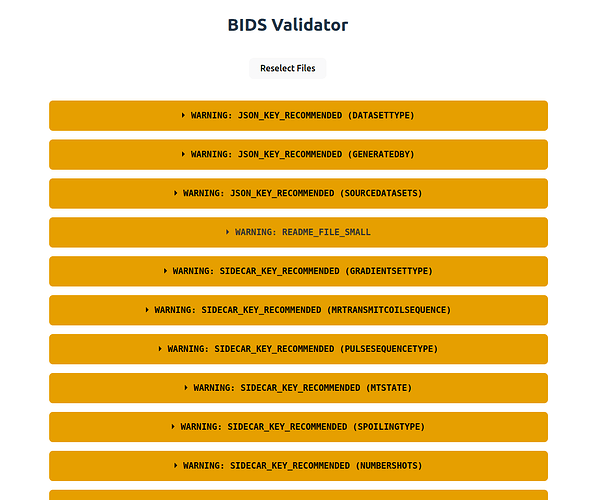
- If I select e.g.
/mnt/data/Investigators/MyStudyI only get warnings. Same happens for eachMyStudy*subfolder, except for one where I see additionally an error related to the4: [ERR] Sub label contain an Illegal Character (...)message that I see in the command line (I see how the file namingsub-155728_T1w_heudiconv323_e1_Eq_1.nii.gzis not BIDS-compliant, but that is not my main issue now I think).
Of note, the validator only detectsMRImodality files: it does not tell me that there is T1, DWI, and field maps.
So I’d like to know what I am missing to properly validate this/these dataset(s). Why do the local and browser versions give different results?
Thanks.