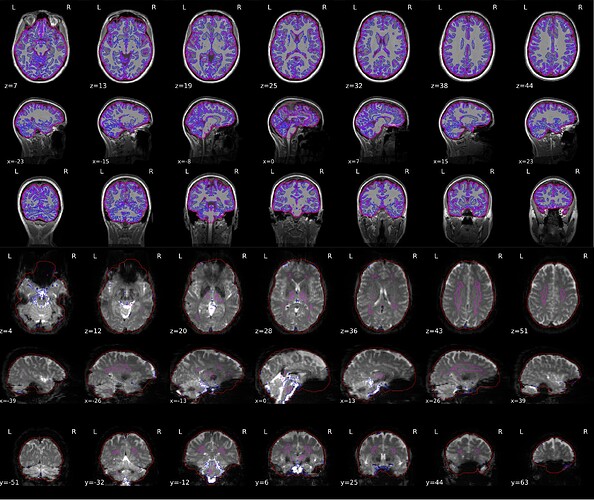
Dear Experts,
we have been running into an odd problem with the BOLD-based brain mask in some participants. This id always located ventral and prefrontally, to varying grades of severity. I appended a picture of the brain masks generated on the T1-weighted and BOLD-based masks below.
I found one post in which this has happened before, which has sadly not been resolved to date.
As I am somewhat new to fMRI analyses, I am sorry if I missed something obvious as to why this might happen.
Thanks and regards,
Sebastian
Command used:
singularity run -B ${DIR_PARENT} /opt/software/fmriprep-20.2.7/fmriprep-20.2.7.sif \
${DIR_BIDS} ${DIR_FMRIPREP} \
participant \
--participant-label $1 \
--skip-bids-validation \
--fs-license-file ${DIR_SCRIPTS}/license.txt \
--fs-no-reconall \
--output-spaces MNI152NLin6Asym:res-2 \
--stop-on-first-crash \
-w ${DIR_FMRIPREP} 2>&1 | tee -a $fmriprep_log
Version:
20.2.7 (We had some issues with our sequences and SDC with newer versions)
Environment:
Singularity container accessed by bash script.
Scanner was a Siemens Prisma Fit 3T.
Screenshots / relevant information:
1 Like
Based on a suggestion by @LoefflerMartin I have created a work-around which at least improves on this issue.
- Use FSL’s BET to extract the brain from the functional data as precisely as possible by playing around with the threshold value
- Use fslmaths -bin to create a binarized mask of the extracted brain
- Use fslmaths -dilM 2-3 times to create a binarized mask of increased size
- Subtract the binarized mask from it’s dilated counterpart and invert the result to get a black outline around the cortex
- Multiply the original input file with the black outline (halo) you created to get nii.gz file that has a very clear outline around the brain
- Use these images as your input for fmriprep
I packaged the whole thing into a bash script that takes input, output and the threshold value for bet as inputs and then does all the bothersome fslmaths stuff for you:
#!/bin/bash
# Check if arguments are provided
if [ $# -lt 3 ]; then
echo "Usage: $0 <input_file> <output_file> <bet_threshold>"
echo "Provide identical input and output to overwrite original file"
exit 1
fi
# Get input and output filenames from command line arguments
input_file=$1
output_file=$2
bet_threshold=$3
echo "Starting bordering for $input_file"
# Run computation
echo "Extracting brain at $bet_threshold threshold"
bet $input_file tmp_brain -f $bet_threshold -g 0
echo "Creating binarized mask"
fslmaths tmp_brain -bin tmp_bin
echo "Dilating binarized mask"
fslmaths tmp_bin -dilM -dilM -dilM tmp_dil
echo "Creating halo mask"
fslmaths tmp_dil -sub tmp_bin tmp_halo_inverted
fslmaths tmp_halo_inverted -mul -1 -add 1 tmp_halo
echo "Subtracting halo mask from original image"
fslmaths $input_file -mul tmp_halo $output_file
# Clean-Up
echo "Removing temporary files"
rm tmp*
# Check if the output file was created successfully
if [ $? -ne 0 ]; then
echo "Error: Failed to create output file."
else
echo "fslmaths command completed successfully."
fi
I appended on of my first results below, I still have to tinker with the size of the outline to possibly get better results, but it’s a start. If you have any comments or suggestions, I would be glad to hear them!
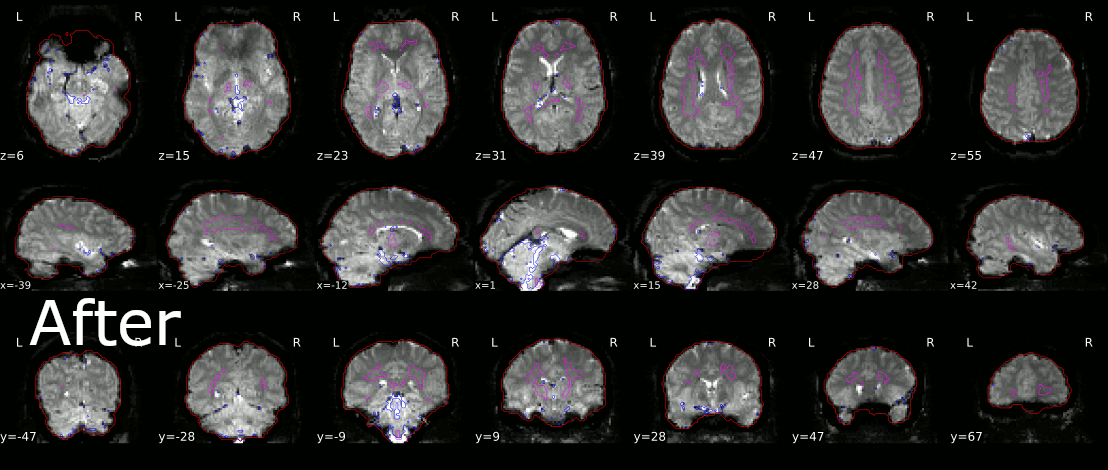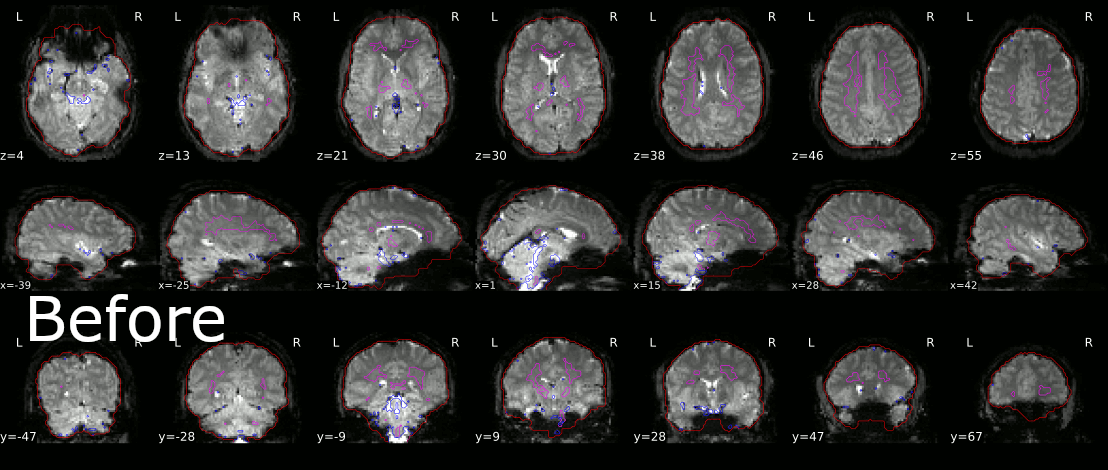
1 Like