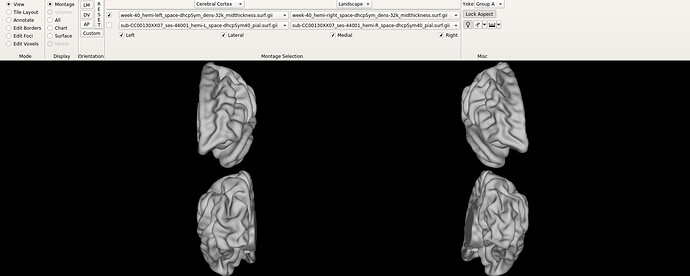
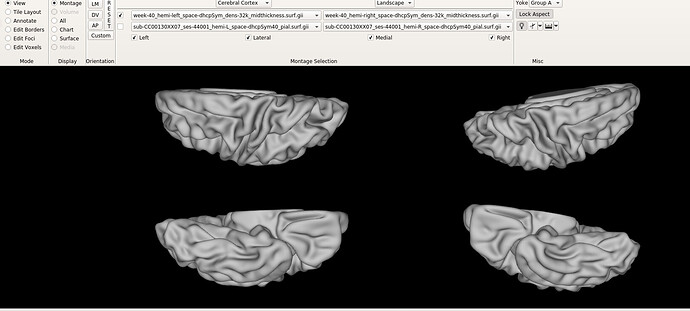
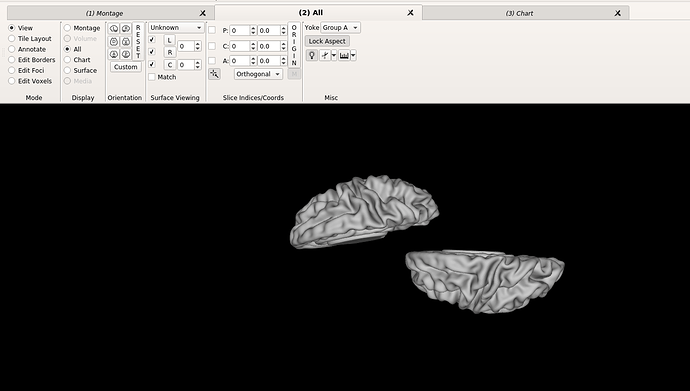
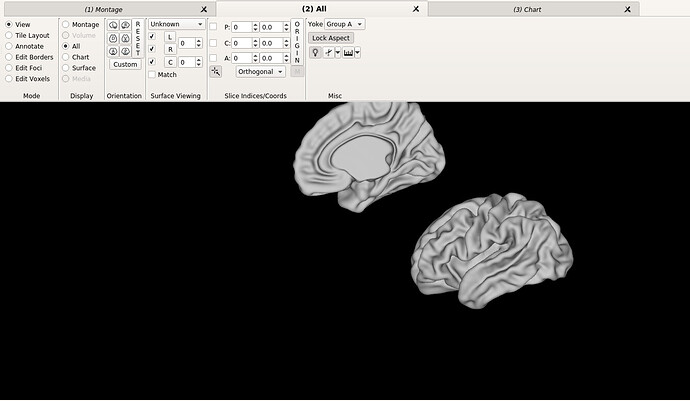
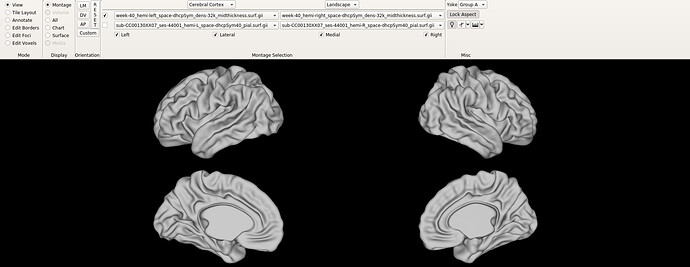
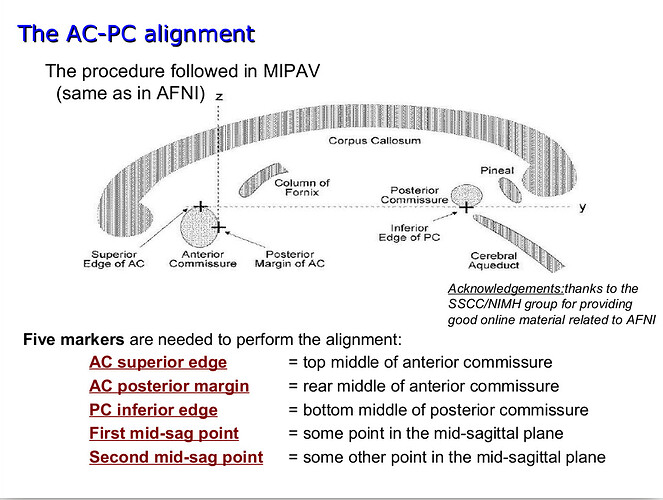
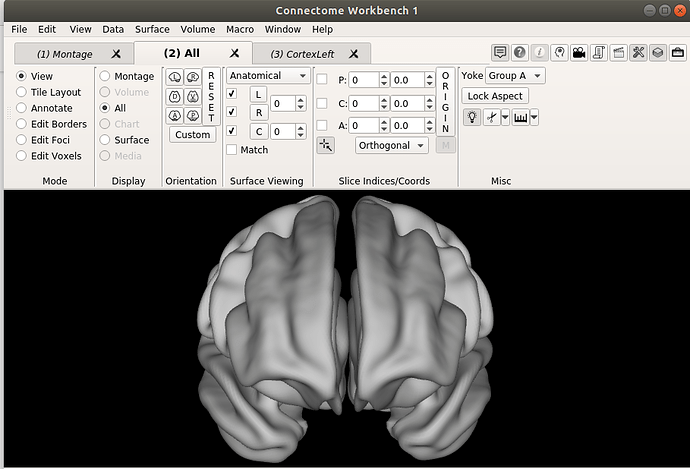
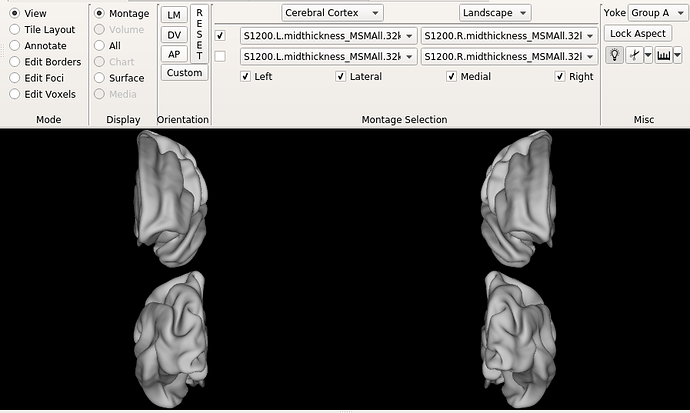
Hello dHCP team,
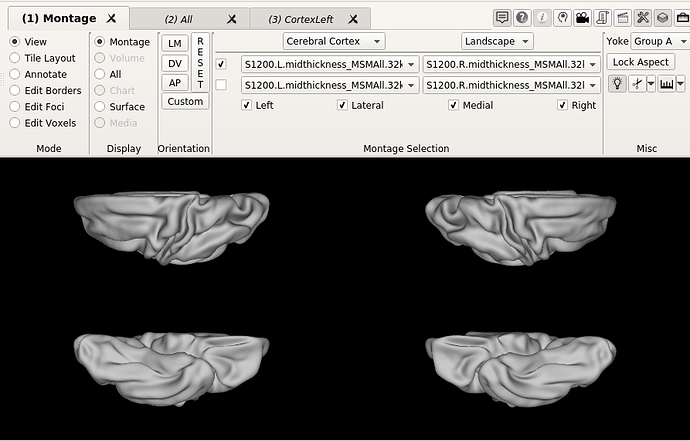
I have a quick question about the alignment of the atlas (Bozek et al., 2018), both the symmetrical and previous versions. On workbench view, the default orientations do not seem to correspond to an alignment where the AC-PC line is an orthonormal y axis, like shown in the following figures:
Is the left hemisphere aligned and then the right one relative to it? Maybe I am posing irrelevant question, since I am not used to work with neonate population, but if that is the case, I wonder why individual subjects seem to have both hemispheres much closely aligned.
Finally, I am not sure if this is related, but when changing to the ‘all’ tab, the right hemisphere drifts completely apart, like shown in the images below.
I am by no means experienced in neuroanatomy, but by simple inspection of the figures below I am not sure the corpus callosum is ACPC aligned? I would appreciate very much any light you could shed on the matter.
Best,
Diego
Hi @diegoder,
I’m not really sure I understand your question. Are you asking about the registration process and template generation? Or are you asking about the orientation of the surfaces in Workbench View?
Best wishes,
Logan
Hi @lzjwilliams,
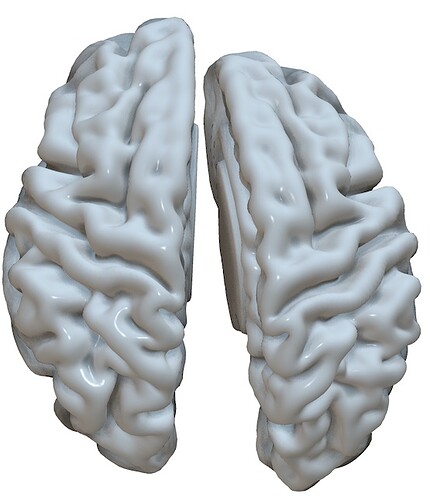
Thank you very much for your answer, I’m sorry the recap paragraph got cut out. My question refers to the generation of the template, and how it orientation looks on workbench view; since the right’s hemisphere especially, looks quite different to the HCP (MNI) atlas attached below.
Thank you!
Ah okay,
So, the original Bozek et al. (2018) atlas should be in the same space as the HCP surface template (fs_LR). The symmetric version of the surface template is in it’s own space (called dhcpSym) - some details about how the template was generated can be found here. Essentially, the left hemisphere was registered to the right, and vice versa. Then the warps of these registrations were averaged to get them into a symmetric space.
Does that answer the question?
Best wishes,
Logan
Thanks for the answer @lzjwilliams,
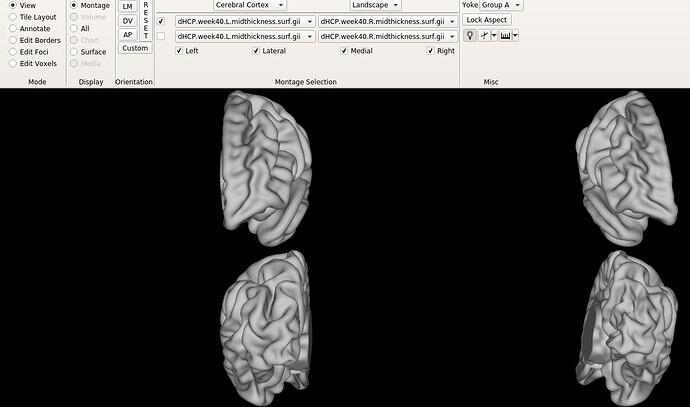
I still don’t understand why the Bozek atlas shows this rotation on the right hemisphere (AP view), as shown in the figure.
Furthermore, when I go to the ‘all’ tab, it is completely empty. Do you think this a problem with my settings on workbench view? Do you get a whole brain surface when viewing the Bozek et al. atlas?
Thank you,
Diego
I don’t have an answer for the rotation of the right hemisphere in the Bozek et al. (2018) atlas, sorry. As for the ‘all’ tab in Workbench, I am able to visualise metric files on the surface. Are you just trying to view the surface?
Best wishes,
Logan
Thank you for the prompt response!
I can’t see the surfaces at all. I’m going to double check wb_view’s settings.
Is the offset in the symmetrical atlas (as shown in the 3rd and 4th figures of original post) also noticeable only in my workbench view? I find it strange that individual subjects and HCP templates look normal.
Best,
Diego
Yes it is. Perhaps @EmmaR could comment on why the right hemisphere is ‘offset’, but I can’t answer any further.
Best wishes,
Logan
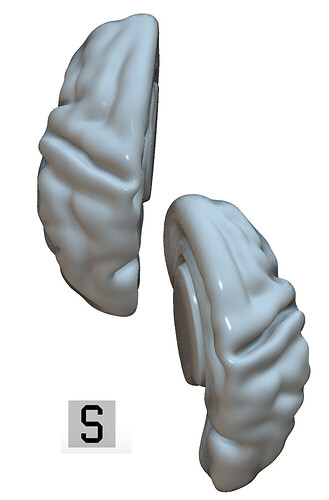
I agree, some of the meshes from the Cortical Surface Atlas (Bozek et al.) are not aligned correctly. Here is a bird’s eye view that shows the odd alignment of dHCP.week42.RH.pial.surf.gii.
Hi @Chris_Rorden & @diegoder,
Having skimmed through Jelena’s paper here, my guess is the reason that they are not aligned is because the neonatal surfaces are initialised by affine registration to an adult surface template (Conte69). Then these neonatal surfaces are averaged to create an initial neonatal template. Then there is an interative process of non-linear registration to this initial template, bringing the neonatal surfaces into better alignment. Then the new registered surfaces are averaged to create an updated template. And the process continues until convergence.
I’ve loaded the HCP average midthickness surfaces on Workbench and see that the medial walls align nicely, compared to the images that you’ve shown here. But I am still unsure how this impacts analysis?
Best wishes,
Logan
@lzjwilliams I suspect you are correct. I do wonder if starting with an age matched template would help, such as one of these.
Hi @lzjwilliams
I had not noticed this as I always use the dhcpSym surfaces.
Can you confirm that this is not a problem for the symmetric surface atlas (dhcpSym)? And if so, can we rigidly align the Bozek surfaces with the dhcpSym to provide a rigid transform to align the left/right hemispheres of the Bozek atlas?
Cheers, Sean
Hi @seanfitz,
The Bozek and dhcpSym atlases have the same orientation. Is that a problem for the vol-to-surface mapping?
Logan
The meshes provided here such as week-28_hemi-left_space-dhcpSym_dens-32k_midthickness.surf.gii are also not correctly aligned. Here is how Surf-ice
Yes, the same alignment “issue” applies to both the Bozek et al atlas and the extended/symmetrised atlas because the symmetric atlas is based on the Bozek et al atlas. But the alignment between the Bozek and symmetric atlases appears almost identical - the symmetric atlas is based on a larger sample size (release 3) compared to Bozek (release 2).
Best wishes,
Logan
No, the vol-to-surface mapping is done with the native surfaces, then resampled to the dhcpSym atlas.