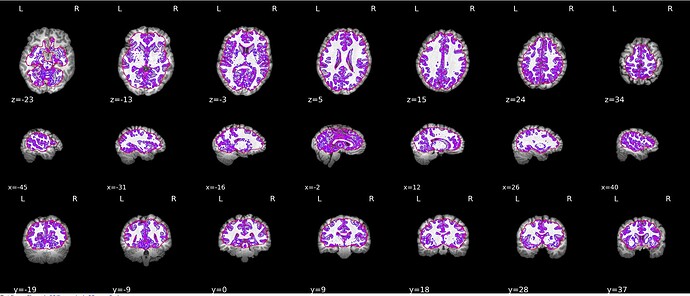
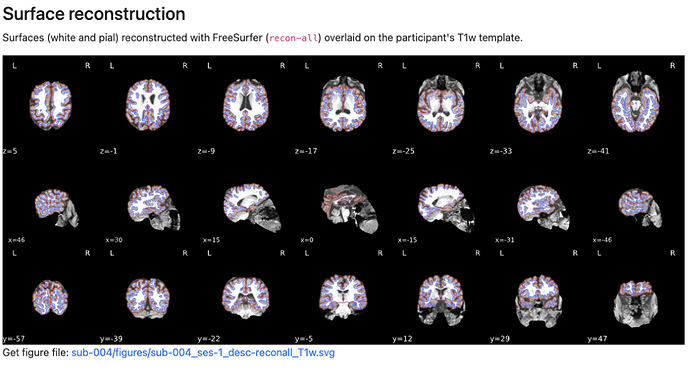
Does anyone had the same issue on brain segmentation ?
Hello and welcome to Neurostars!
It would help if you could provide the following:
- What version of fmriprep/smriprep you are using.
- Did it complete without errors? If there was an error, what is full traceback?
- Is this error subject specific?
- Your full command.
- Anything particularly noteworthy about your data organization (e.g. multiple T1s)?
Best,
Steven
If you’re passing pre-skullstripped T1w images, you’ll need to use --skull-strip-t1w skip. Unfortunately, auto-detecting skullstripped images is not as trivial as we’d hoped and we’ve left it to users to specify.
Hi steven thank for the reply,
As Chris Markiewicz pointed out it is a skul stripping issue , may data are alreday skull stripped and the --skull-strip-t1w option is by default at force.
thanks you so much and next time I’ll post a full description of my issue ![]()
That’s it Chris, I’m testing with skull stripping off. I’m quite sure it will fix the problem.
Thank you so much Chris
Hello!
I am using fmriprep 21.0.1 in a docker.
I have one subject where skull stripping doesn’t seem to be as clean as I would like it to be (see below image). fmriprep completes without any errors but the reconstruction looks like:
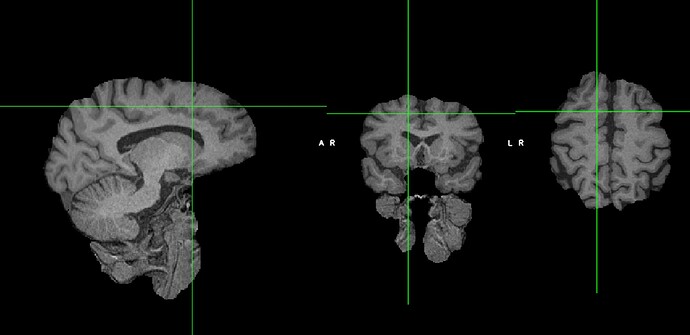
I skull stripped via the FSL GUI using BET brain extraction and when I view that skull-stripped image it looks like this:
(so it looks like the fmriprep reconstructed image)
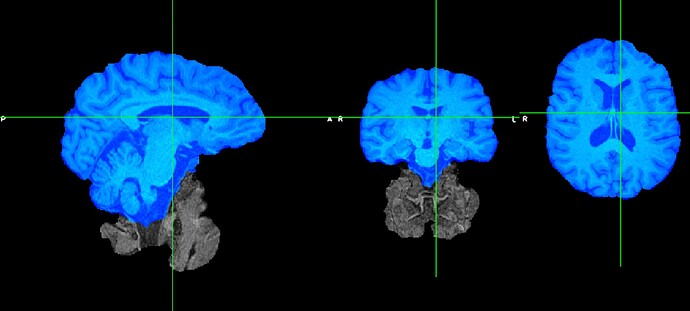
And then I run another brain extraction on that 1st round of BET extraction and it’s cleaned up (below I took a screenshot of the 2nd extracted brain in blue overlaying the first extracted brain).
So, I think I would like to use this second pre-skullstripped T1 image but I’m not sure where/which folder to put this image into so that fmriprep knows to use it. Any insight/suggestions would be helpful! Let me know if I can provide clarifying information.
Thank you!
edit: Oh, if it’s important, this is a brain that has lesions so I’ve included the lesion_roi.nii.gz image in their Nifti/anat folder along with their T1w, T2w, and FLAIR images.
Hi @fareshte,
Please open a new issue under the software support category, making sure to fill out information in the prepopulated template. Also try upgrading to the most recent release (23.0.1 at this time).
Best,
Steven