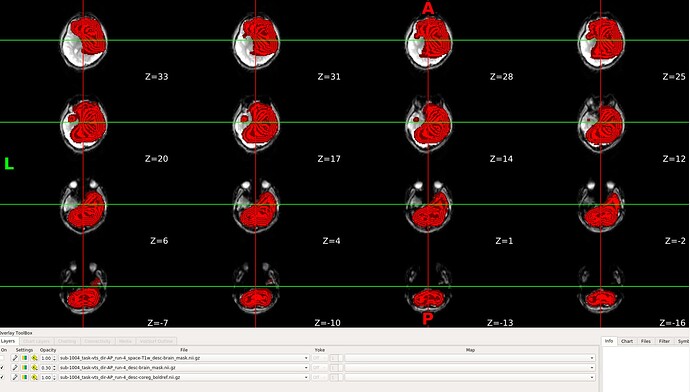
I’m having some issue with the brain mask on 1 of 4 runs - the other runs are all fine. The mask is badly distorted as seen here in red overlay on a boldref:
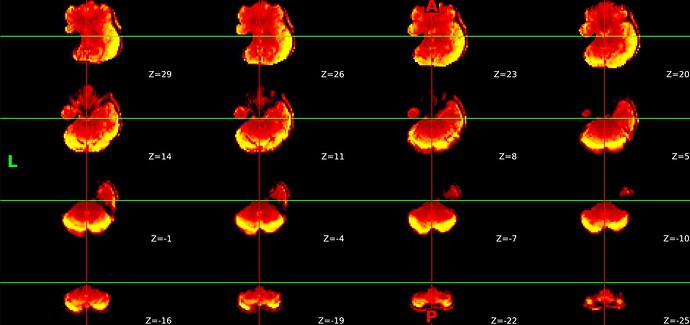
It appears this is having downstream effects on the preprocessed output. Here’s the T1w space preproc bold:
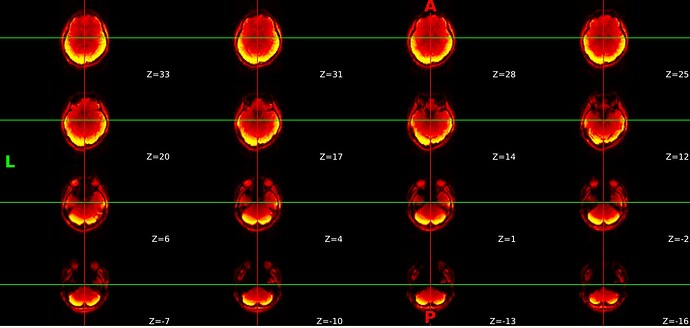
However, I have multi-echo data and my single echo preproc files are fine:
So while I can still run TEDANA on the single echo files, I can’t apply the MNI registrations from fmriprep to the tedana output as they were based on the distorted files. I followed a previous suggestion to create a mask from the union of CSF, WM, and GM probseg maps, but I still have the problem of not having a good reference image in MNI space to use for antsApplyTransforms.
Does anyone have any suggestions for a workaround that doesn’t involve manually running ants registration?