Hello everyone,
We have a 7T dataset with anatomical (MPRAGE), topup and functional images (3DEPIs) for all our 19 subjects. They have been preprocessed with #fmriprep 20.0.6 . We are encountering some troubles in the anatomical processing.
-
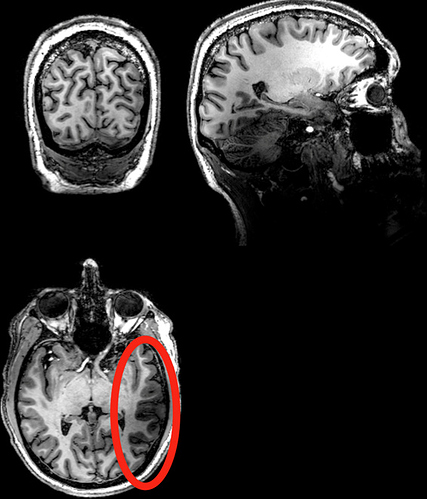
fMRIprep is outputting some less than ideal anatomical brain masks in some participants. This is because of the right occipital lobe being a bit darker due to some problems we had with the scanner (this is consistent throughout all of the data of those participants). These parts of the cortex are being left out of the anatomical brain mask, which we observe clearly in the fmriprep reports.
As a result, surface reconstruction fails of course, but this is not our main concern at the moment.
Despite this problem, the normalization of the anatomy to MNI space and the coregistration with the functional images looks ok.
It is unclear to us however in what other steps the anatomical brain mask is used.
As far as we understand, it is not used in motion correction or susceptibility distortion correction (with topups) of the functional images.
However, we are worried that the anatomical brain mask is in some way used during the normalization of the functional images. We want to make sure no information is being cut out of the functional scans before they are transformed to MNI space…
Is it used during functional normalization? We cannot clearly find an answer to this question.
If anyone can help out with this worry, we’d be very grateful!
-
In addition, one subject had a MP2RAGE anatomy collected instead of MPRAGE, and also here the anatomical brain mask failed for some reason; almost all of the grey matter is left out of the mask, which messes up the anatomical normalization to MNI space. If you would have any advice on how to handle this specific subject so that we do not have to exclude it in our study, that would be great.
Thank you!
Can you post examples?
Well, this means that your data might be usable after all. But definitely, use a lot of caution.
The anatomical brain mask is mostly used in normalization to standard space, and for certain T1w preprocessing steps. I would very carefully check on the WM-GM surface extracted by FreeSurfer, because that will mostly drive the co-registration with the fMRI runs - although you mention that also looks good.
The problems will arrive later, if surfaces are wrong, you’ll immediately discard CIFTI outputs or surface analyses. And building confidence in the spatial normalization (to use any sort of atlas/parcellation) will also be a hard tasks.
Yes, to normalize the BOLD data into standard space you need to apply concatenated the transforms from standard-space to T1w and from T1w to BOLD.
All said above stands for any pipeline, not just fMRIPrep. If your data have prominent issues, they can potentially introduce biases in the final result. So these data must be handled with great care (and report transparently all the little tweaks your tooling required and all the quality control checkpoints you set in place)
That is a longstanding issue (https://github.com/nipreps/smriprep/issues/18) for which I can’t foresee any prompt resolution. You could brain-extract this image on your own and tell fMRIPrep the image is skull-stripped.
BTW a fairer title to this thread would be: Data with pretty serious quality issues seem to be reasonably digested by fMRIPrep - can I trust the results?
Thank you for your answer and your help!
As you mention, the grey and white matter surface results aren’t much good. However, even though this drives the co-registration with the functional scans as you say, it seemed co-registration in all the subjects with this issue looked alright aka locations in both the anatomical and functional scans are in alignment.
If we would manually correct the surfaces in FreeSurfer (if that would be feasible) and reconstruct them again, then would we able to normalize the surface and be able to use an atlas/parcellation? Because we would like to use a surface-based atlas.
We’re unsure about the practical details of the co-registration and functional normalization steps; in particular what the implications of certain transformations could be for the data.
What I want to be sure about for both the co-registration and the functional normalization, and where I still feel confused: are the parts of the brain, that are left out of the anatomical brain mask, also cut out of the functional images during co-registration or its normalization and then filled up again/replaced with BOLD signal that doesn’t use the BOLD signal that is originally present in those cut-out areas in the functional scans? That would be detrimental since we are looking for experimental effects in those regions that are being cut out in the anatomical brain mask, so we don’t want them cut out of the functional scans and replaced with data that doesn’t reflect the original data/effects present in those areas.
I’m unsure about how to phrase my question clearly, I apologize for any confusions.
As for the MP2RAGE issue, thank you for the link and the advice as to what we can try!
We will definitely handle this data with great care, and thanks again for your help.
Do you have any advice for a tool or a certain step we could perform on the data before sending it through fMRIprep, to avoid the issue we’re facing? E.g. skull-stripping or some kind of inhomogeneity correction?
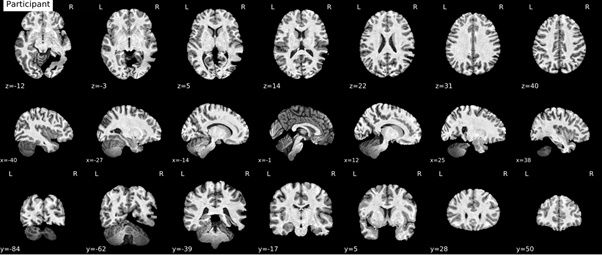
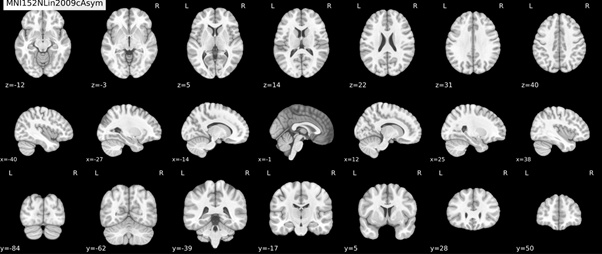
Here are examples of the raw data as well as the brain mask and the normalization:
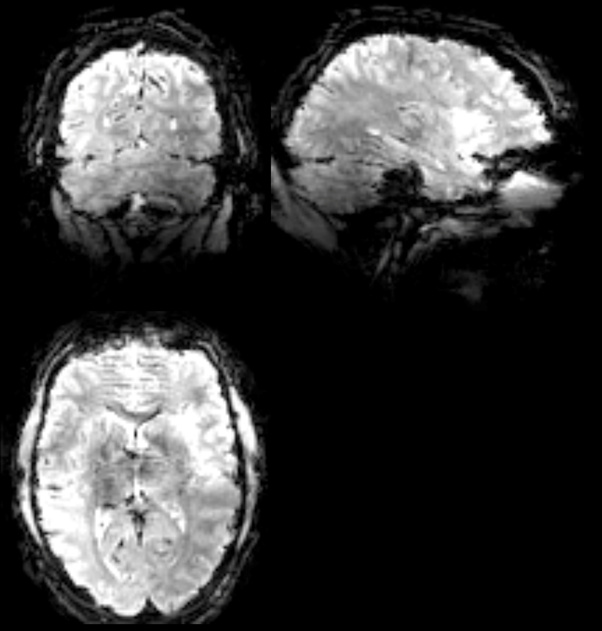
Examples of raw anatomy and functional data that show the darker region:
anatomical brain mask:As an example of the anatomical brain mask issue. It’s most clearly visible in y=-54 and y=-33.
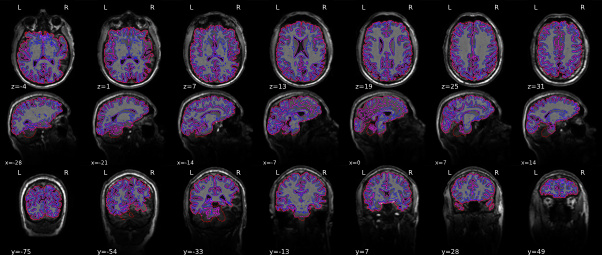
anatomical normalization: