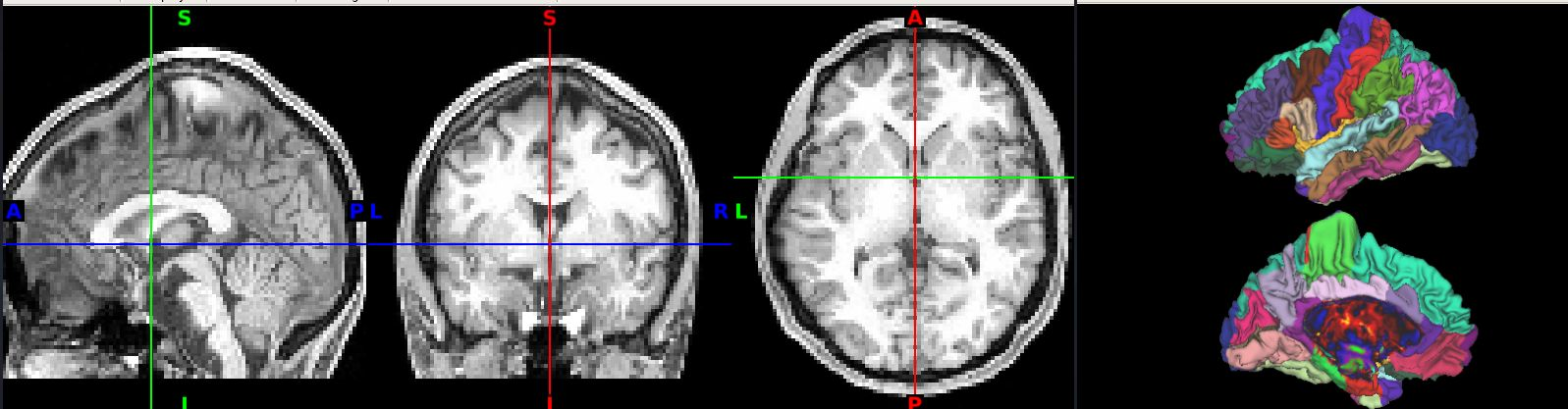
The original post is here (T1w anatomical normalization issue · Issue #171 · edickie/ciftify · GitHub), but I am reposting here on Neurostars to see if someone has the same issue.
Ciftify completes without error on the subjects I’ve tried so far, but I’m getting what looks to be bad Ciftify-based MNI normalizations for the fmriprep preprocessed T1w images. I believe where MNI normalization occurs is in the “Registering T1wImage to MNI template using FSL FNIRT” step of ciftify_recon_all.
The output from a subject from our lab shows the issue (note the distortion along the top of the brain, particularly in the pre/post central regions)
Have anyone seen this before? Is there a way to prevent this or improve the normalization?
Does the T1w normalization issue impact the analysis of the surface fMRI data produced by Ciftify?
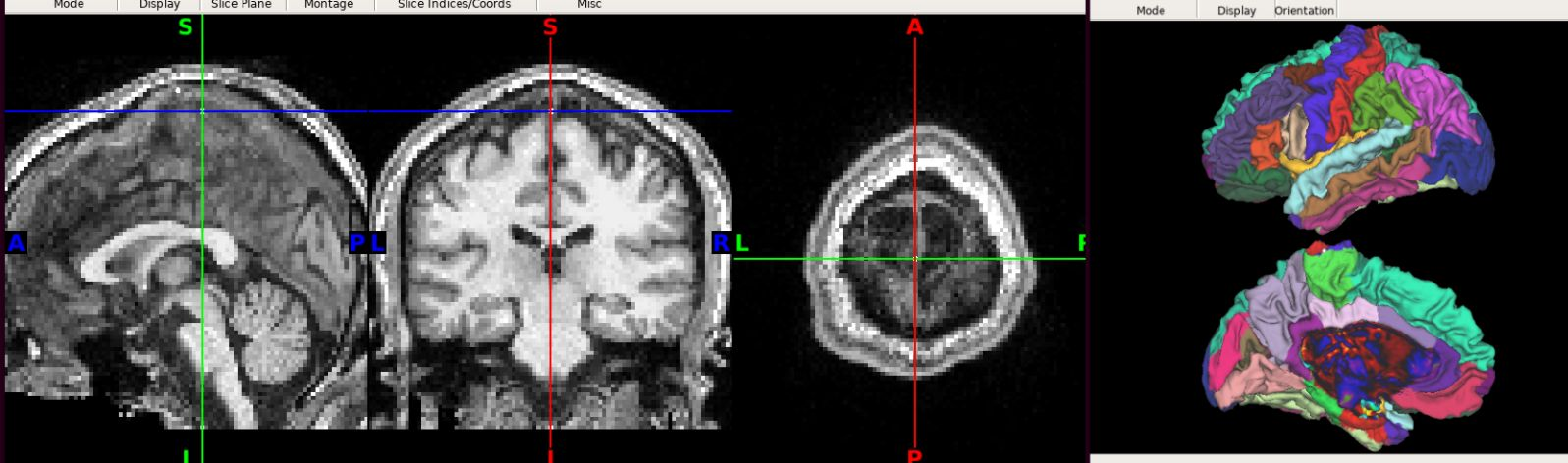
Also, here is was I got for one the Ciftify example subjects (sub-50004 from the tutorial). This subject also shows some slight deformity in the pre/post central gyral regions, although not as extreme as in the above case.
Here are my settings …
System Info:
OS: Linux
Hostname: mica
Release: 3.10.0-862.3.2.el7.x86_64
Version: #1 SMP Mon May 21 23:36:36 UTC 2018
Machine: x86_64
ciftify:
Version: 2.3.3
wb_command:
Path: /onrc/home/pipeline/data/pipelineb/home/Chyatt/onrc/data/Apps/workbench/bin_rh_linux64/wb_command
Version: 1.5.0
Commit Date: 2021-02-16 13:46:47 -0600
Operating System: Linux
freesurfer:
Path: /opt/freesurfer-6.0/bin
Build Stamp: freesurfer-Linux-centos6_x86_64-stable-pub-v6.0.0-2beb96c
FSL:
Path: /opt/fsl/fsl-6.0.3
Version: 6.0.3:b862cdd5
—### End of Environment Settings ###—
Issue also occurs with FSL v5.0.11.