Dear all,
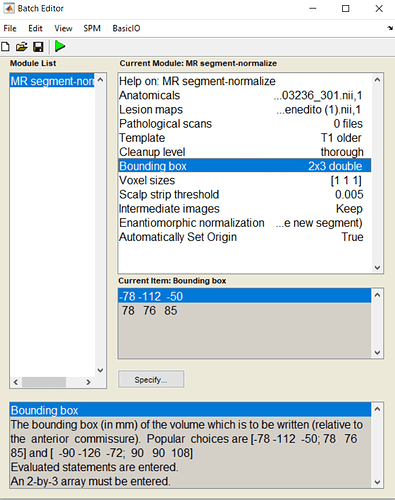
When I tried to run the ‘MR segment-normalize’ module of the Clinical Toolbox, I got the following error:
12-May-2021 16:29:03 - Running ‘MR segment-normalize’
12-May-2021 16:29:03 - Failed ‘MR segment-normalize’
Undefined function ‘clinical_mrnormseg_job’ for input arguments of type ‘struct’.
In file “C:\Users\Neuroimager\Desktop\spm12\toolbox\Clinical-master\tbx_cfg_clinical.m” (???), function “clinical_local_mrnormseg” at line 319.
The following modules did not run:
Failed: MR segment-normalize
For more details of the information entered:
I would appreciate it very much if anyone could help me on this issue.
Thanks so much,
Luan.
If you download from Github, make sure you rename the folder Clinical-master to Clinical, e.g. remove the branch name. In future, I guess I can make Clinical a folder inside the repository to avoid confusion.
1 Like
Thank you so much for your help and attention! 
I’ m a beginner in this area, and your help has been invaluable.
Dear all,
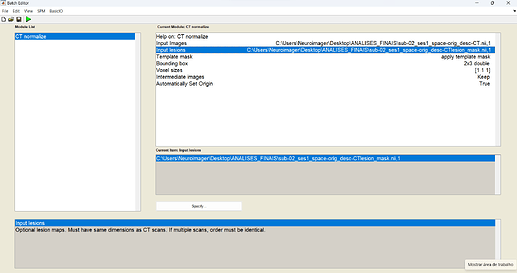
When I tried to run the ‘CT normalize’ module of the Clinical Toolbox, I got the following error:
SPM present working directory:
C:\Users\Neuroimager\Desktop\ANALISES_FINAIS
SPM12: spm_image (v7573) 10:58:04 - 19/12/2023
========================================================================
------------------------------------------------------------------------
19-Dec-2023 10:58:52 - Running job #1
------------------------------------------------------------------------
19-Dec-2023 10:58:52 - Running 'CT normalize'
CT normalization version 7/7/2016
clinical_fix_ge_ct skipped: This image does not have unusual image intensities: C:\Users\Neuroimager\Desktop\ANALISES_FINAIS\sub-02_ses1_space-orig_desc-CT.nii,1.
C:\Users\Neuroimager\Desktop\ANALISES_FINAIS\sub-02_ses1_space-orig_desc-CT.nii center of brightness differs from current origin by -0x-19x9mm in X Y Z dimensions
------------------------------------------------------------------------
19-Dec-2023 10:58:53 - Running job #2
------------------------------------------------------------------------
19-Dec-2023 10:58:53 - Running 'Coregister: Estimate'
SPM12: spm_coreg (v7320) 10:58:53 - 19/12/2023
========================================================================
Printing 'Graphics' figure to:
C:\Users\Neuroimager\Desktop\ANALISES_FINAIS\spm_2023Dec19.ps
Completed : 10:59:24 - 19/12/2023
19-Dec-2023 10:59:24 - Done 'Coregister: Estimate'
19-Dec-2023 10:59:24 - Done
C:\Users\Neuroimager\Desktop\ANALISES_FINAIS\sub-02_ses1_space-orig_desc-CT.nii intensity range: 2967
Ignore QFORM0 warning if reported next
------------------------------------------------------------------------
19-Dec-2023 10:59:27 - Running job #2
------------------------------------------------------------------------
19-Dec-2023 10:59:27 - Running 'Image Calculator'
SPM12: spm_imcalc (v6961) 10:59:27 - 19/12/2023
========================================================================
ImCalc Image: C:\Users\Neuroimager\Desktop\ANALISES_FINAIS\dxsub-02_ses1_space-orig_desc-CTlesion_mask.nii
19-Dec-2023 10:59:27 - Done 'Image Calculator'
19-Dec-2023 10:59:27 - Done
clinical_smoothmask: C:\Users\Neuroimager\Desktop\ANALISES_FINAIS\xsub-02_ses1_space-orig_desc-CTlesion_mask.nii had 47043 voxels >0.001000
masking C:\Users\Neuroimager\Desktop\ANALISES_FINAIS\sub-02_ses1_space-orig_desc-CT.nii with C:\Users\Neuroimager\Desktop\ANALISES_FINAIS\ssub-02_ses1_space-orig_desc-CTlesion_mask.nii using template C:\Users\Neuroimager\Desktop\Backup_LuanAguiar\Pacotes_Neuroimagem\spm12\toolbox\Clinical\scct.nii.
Conversion Normalise:EstWrite -> Old Normalise:EstWrite
------------------------------------------------------------------------
19-Dec-2023 10:59:28 - Running job #2
------------------------------------------------------------------------
19-Dec-2023 10:59:28 - Running 'Old Normalise: Estimate & Write'
Smoothing by 0 & 8mm..
Coarse Affine Registration..
Fine Affine Registration..
19-Dec-2023 10:59:29 - Failed 'Old Normalise: Estimate & Write'
Error using spm_affreg
Source and its weighting image must have same orientation.
In file "C:\Users\Neuroimager\Desktop\Backup_LuanAguiar\Pacotes_Neuroimagem\spm12\toolbox\OldNorm\spm_affreg.m" (v7377), function "spm_affreg" at line 66.
In file "C:\Users\Neuroimager\Desktop\Backup_LuanAguiar\Pacotes_Neuroimagem\spm12\toolbox\OldNorm\spm_normalise.m" (v5745), function "spm_normalise" at line 182.
In file "C:\Users\Neuroimager\Desktop\Backup_LuanAguiar\Pacotes_Neuroimagem\spm12\toolbox\OldNorm\spm_run_normalise.m" (v4873), function "spm_run_normalise" at line 33.
The following modules did not run:
Failed: Old Normalise: Estimate & Write
19-Dec-2023 10:59:29 - Failed 'CT normalize'
Error using MATLABbatch system
Job execution failed. The full log of this run can be found in MATLAB command window, starting with the lines (look for the line showing the exact #job as displayed in this error message)
------------------
Running job #2
------------------
The following modules did not run:
Failed: CT normalize
For more details:
Previously, I believed that it could be the step I performed before trying to perform normalization, which would be “reorientation” (defining the origin ‘anterior commissure’ and the orientation of the patient’s image manually in the SPM[Display]).
I would appreciate it very much if anyone could help me on this issue.
Thanks so much,
Luan.