Summary of what happened:
Hi all!
I am trying to build an external custom BIDS atlas to import within xcp_d to extract custom connectivity matrices. To do so, I need to create a dlabel.nii file containing my left/right cortical labels and subcortical labels. For the cortical labels, I have two gifti files: child_164k_L.label.gii and child_164k_R.label.gii. For the subcortical label, I have a single nifti file: BN_child_subcortical.nii.gz. The final atlas needs to be in 32k space. For now, here are the steps I used (see code below):
- Add label data to the subcortical volume
- Combine both hemispheres and subcortical data into a single
dlabel.niifile in 164k space. - Resample the
dlabel.niifile in the 32k space.
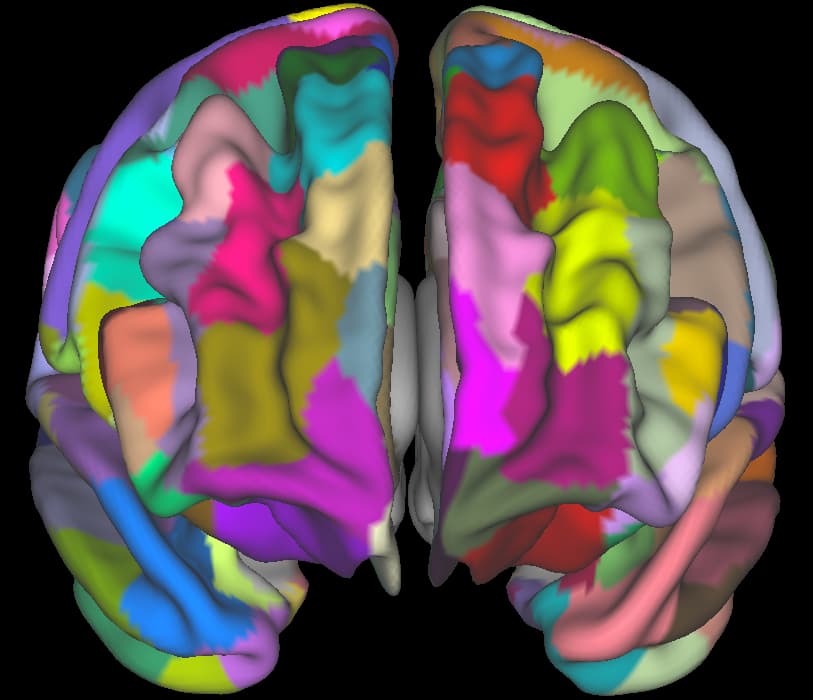
Everything seems to run fine (no specific log), and I can visualize my cortical labels in wb_view (unsure how to visualize subcortical labels though):
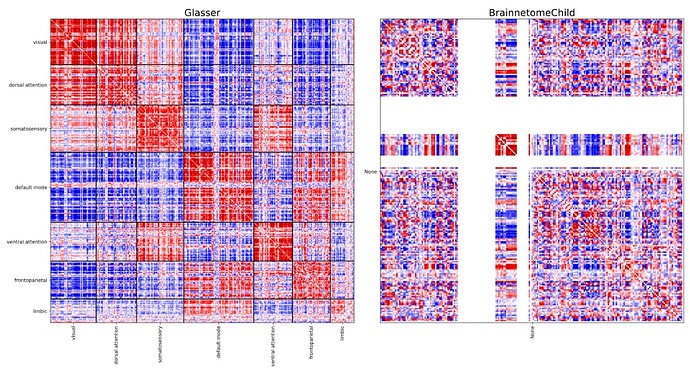
However, when I run xcp_d, I obtain FC for every cortical pairs, but only N/A for the subcortical regions.
I’ve searched other topics and resources, but every solutions did not seem to work in my specific use case. Anyone has a suggestion I could try? I can share the required files if necessary.
Command used (and if a helper script was used, a link to the helper script or the command generated):
# Create a cifti header in the nifit file.
wb_command -volume-label-import ${WDIR}/temp/BN_child_subcortical.nii.gz ${WDIR}/BN_child_parcel_labels.txt ${WDIR}/temp/BN_child_subcortical.nii -drop-unused-labels
# Create the 164k version of the dense nifti files.
wb_command -cifti-create-label ${WDIR}/temp/atlas-BrainnetomeChild_space-fsLR_den-164k_dseg.dlabel.nii \
-volume ${WDIR}/temp/BN_child_subcortical.nii ${WDIR}/temp/BN_child_subcortical.nii \
-left-label ${LEFT_ATLAS} \
-right-label ${RIGHT_ATLAS}
# Resample the 164k dense nifti files to 32k using fsaverage spheres.
wb_command -cifti-resample ${WDIR}/temp/atlas-BrainnetomeChild_space-fsLR_den-164k_dseg.dlabel.nii COLUMN \
${REFERENCE} COLUMN BARYCENTRIC ENCLOSING_VOXEL temp/atlas-BrainnetomeChild_space-fsLR_den-32k_dseg.dlabel.nii \
-left-spheres ${TEMPLATEFLOW_HOME}/tpl-fsLR/tpl-fsLR_space-fsaverage_hemi-L_den-164k_sphere.surf.gii ${TEMPLATEFLOW_HOME}/tpl-fsLR/tpl-fsLR_space-fsaverage_hemi-L_den-32k_sphere.surf.gii \
-right-spheres ${TEMPLATEFLOW_HOME}/tpl-fsLR/tpl-fsLR_space-fsaverage_hemi-R_den-164k_sphere.surf.gii ${TEMPLATEFLOW_HOME}/tpl-fsLR/tpl-fsLR_space-fsaverage_hemi-R_den-32k_sphere.surf.gii
Version:
Connectome Workbench
Type: Command Line Application
Version: 2.0.1
Qt Compiled Version: 6.2.3
Qt Runtime Version: 6.2.3
Commit: 150de12f4f4b94b39bec6d9133ad2e7019d2d3ef
Commit Date: 2024-10-15 17:38:41 -0500
Compiler: c++ (/Applications/Xcode.app/Contents/Developer/Toolchains/XcodeDefault.xctoolchain/usr/bin)
Compiler Version: 13.0.0.13000029
Compiled Debug: NO
Operating System: Apple OSX
Compiled with OpenMP: YES
Environment (Docker, Singularity / Apptainer, custom installation):
Source install.
Data formatted according to a validatable standard? Please provide the output of the validator:
#Not relevant here
Relevant log outputs (up to 20 lines):
# No logs outputs.