Thanks @dowdlelt!
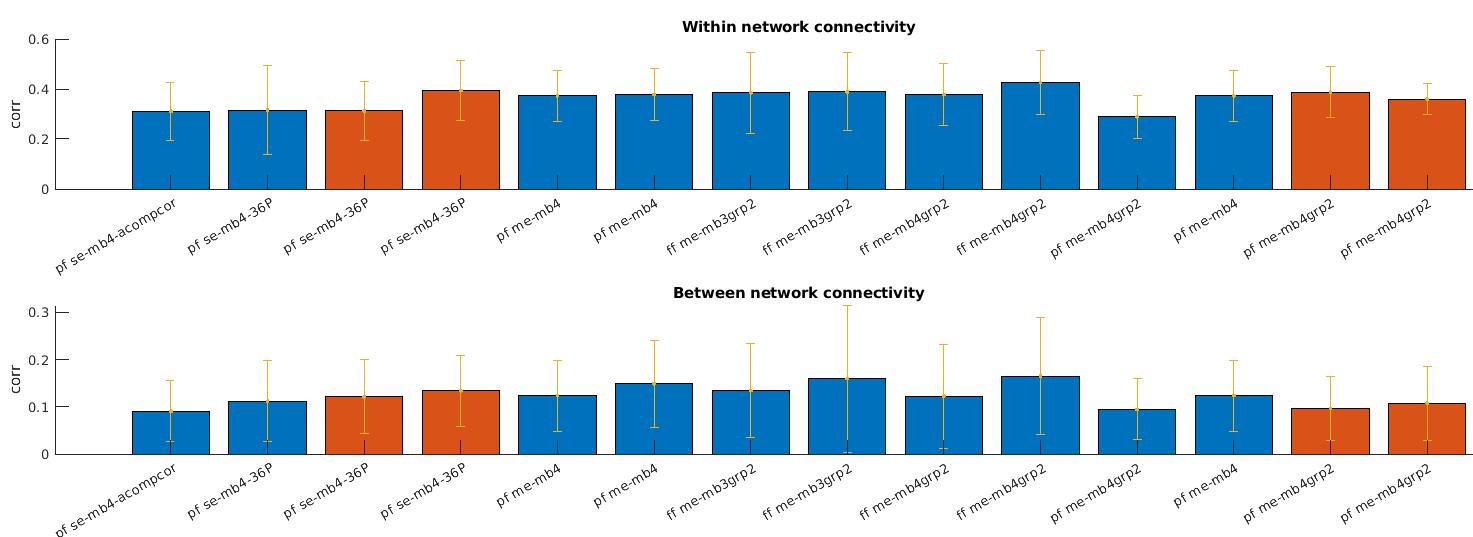
- To compare the different protocols I calculated the within-network connectivity (should be high) and between-network connectivity (presumably, should be low) for each protocol, after running tedana+xcpd with aCompCor regressors. For the single-echo scans I only applied xcpd. The two colore are two different subjects, and the single-echo scans are the first 4 bars.
se - single echo
me - multi-echo
mb - multi-band
grp - grappa
pf - partial Fourier
ff - full Fourier
Unfortunately, based only on this sample, it doesn’t seem like the mean+std of these measures improves much. Perhaps for the orange subject it does improve slightly between single-echo and multi-echo scans. Would you expect greater effects? Or is it too hard to say based on so few scans?
- I didn’t think about the time SBRef takes. You meant that noise might still leak into the data via motion during the SBRef scan, correct?
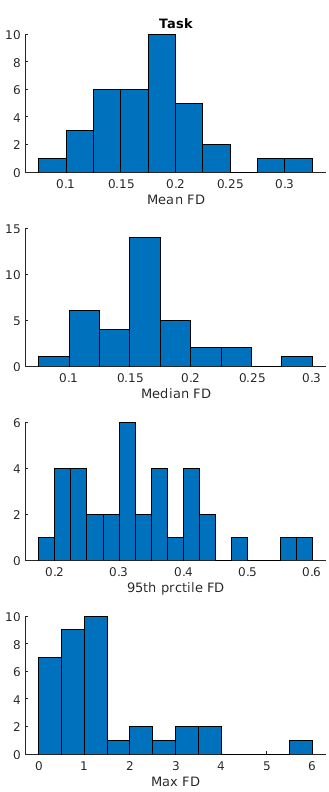
I hope this is not off-topic, but I don’t know what you consider to be low-motion for subjects. Here are some histograms shwing subject counts of several summary statistic of Framewise Displacement: in our data:
While the maxiam DF can get quite large, the median is always below 0.3mm.
Would you consider these to be low-motion?
Thanks for sticking here with me ![]()