Dear community members,
I have recently started working with Multi-Band Multi-Echo (MBME) data in a pipeline including fMRIprep and TEDANA (see details of scan protocols below). To test the pipeline, I collected fixation (“resting state”) data with two protocols.
I used fMRIprep 23.2.0, which outputs the single-echos after head motion correction + suceptibility distortion correctoin (no slice-timing correction due to short TR), and subsequently ran TEDANA (23.0.1) with the default tedpca=‘ica’ option.
As I have no experience in analyzing ME data, I would love to get your insight in order to choose the best protocol. My concerns are the following (I hope it’s okay they’re all in a single post):
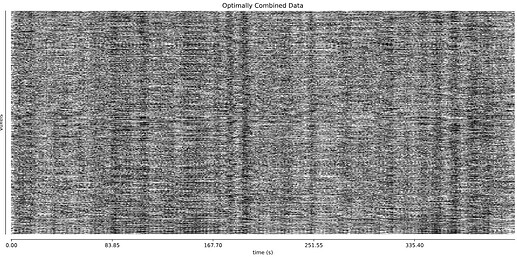
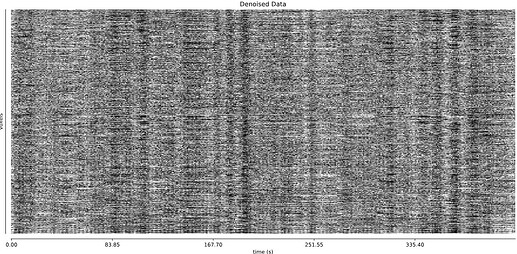
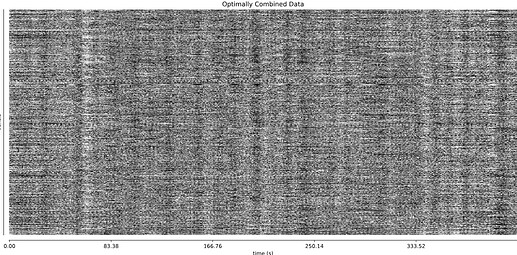
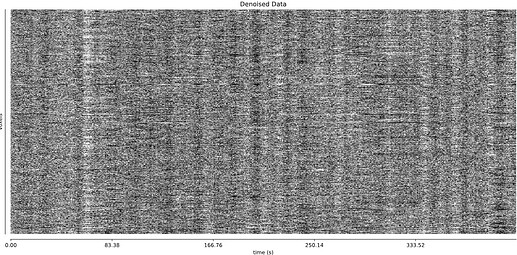
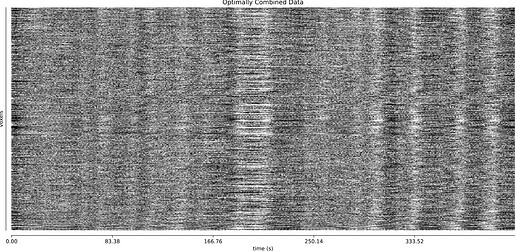
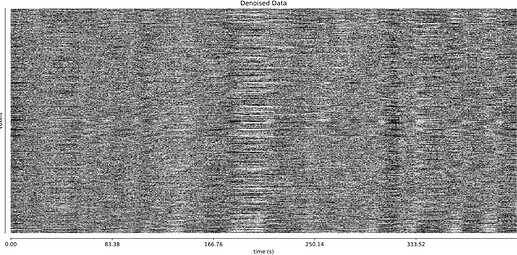
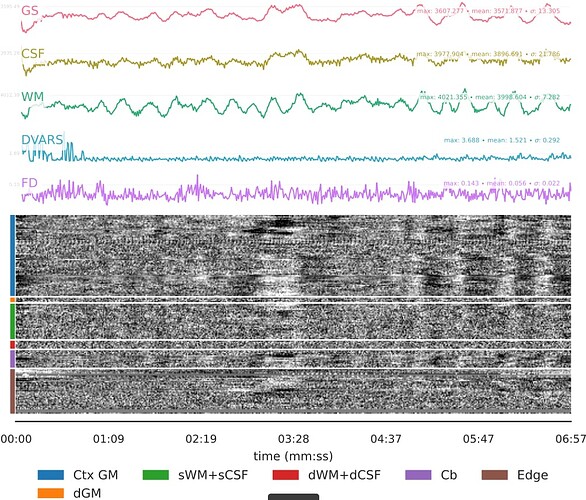
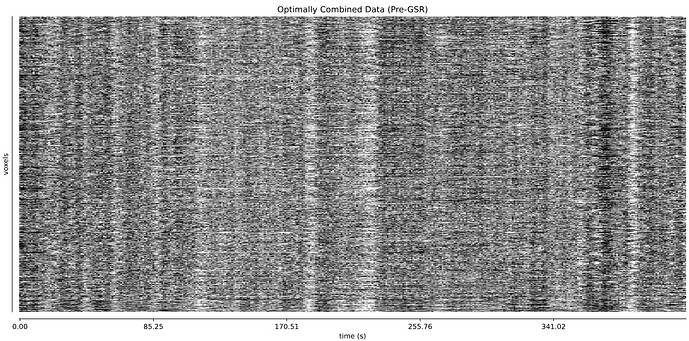
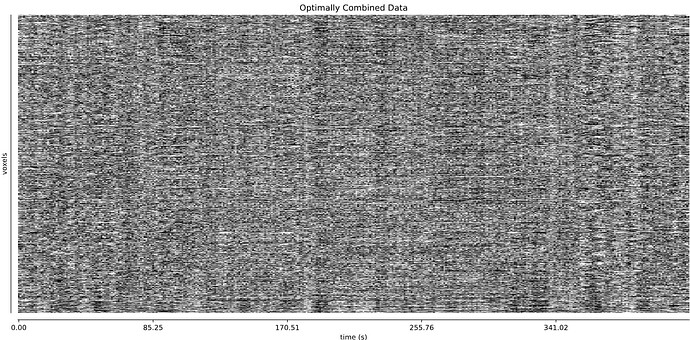
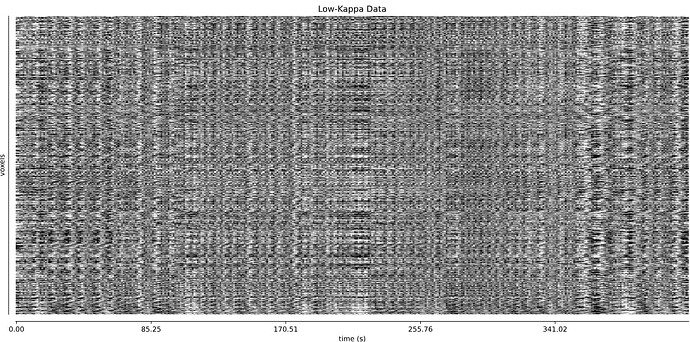
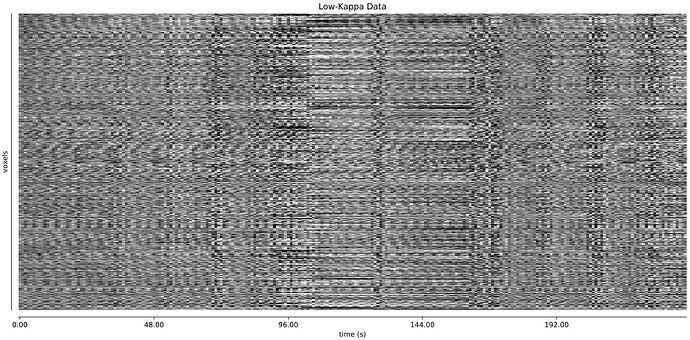
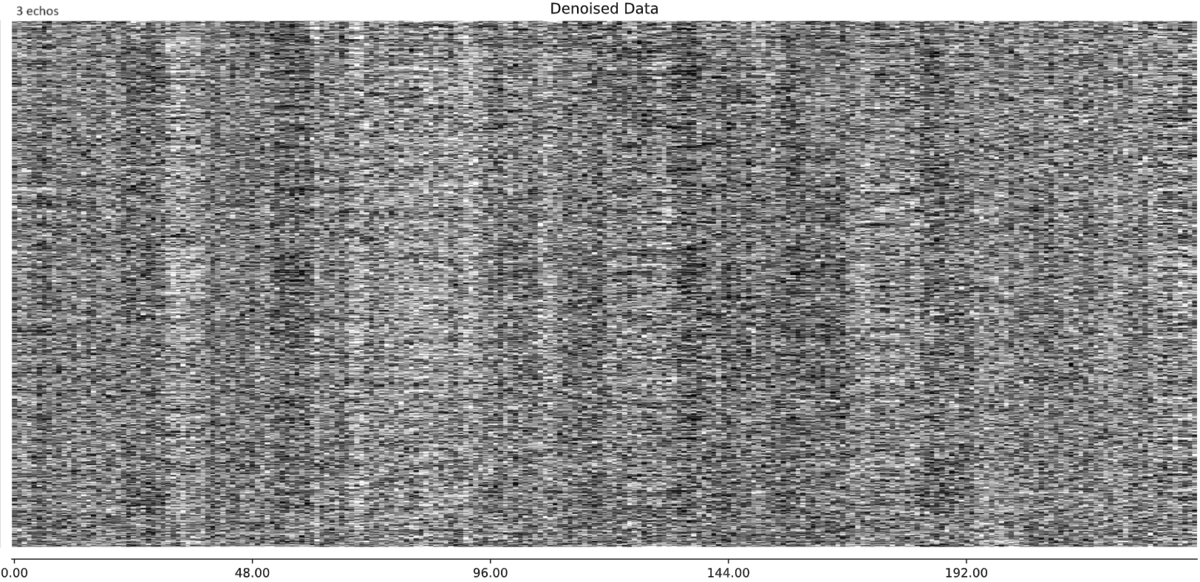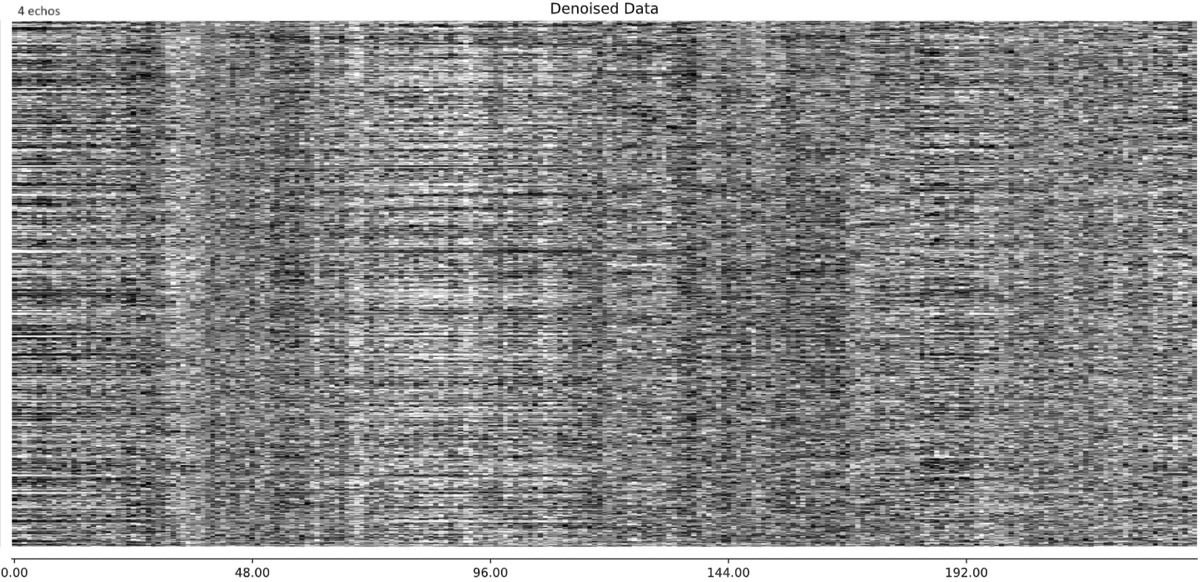
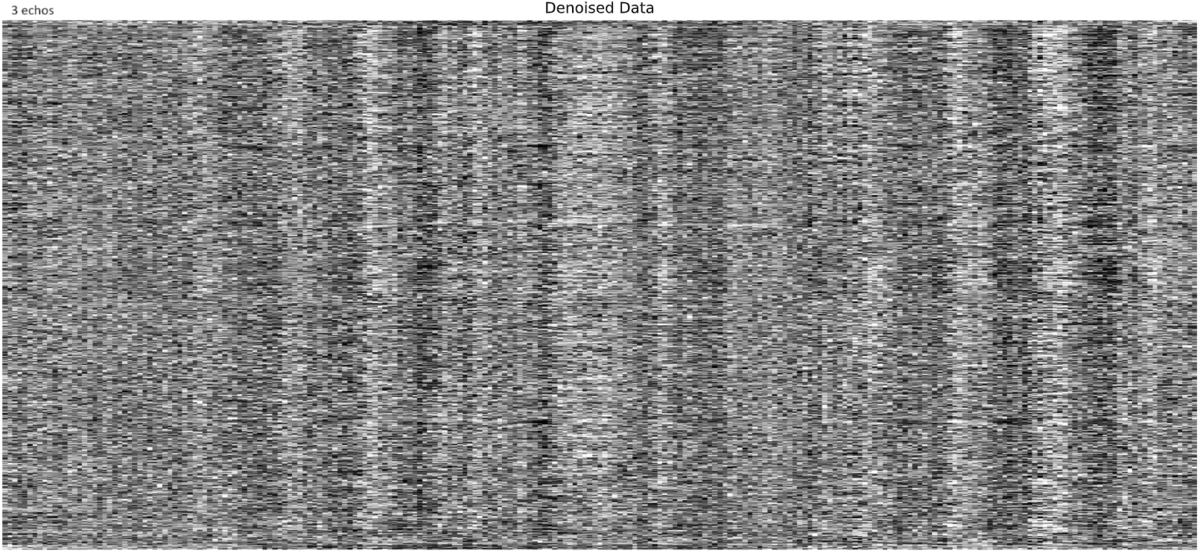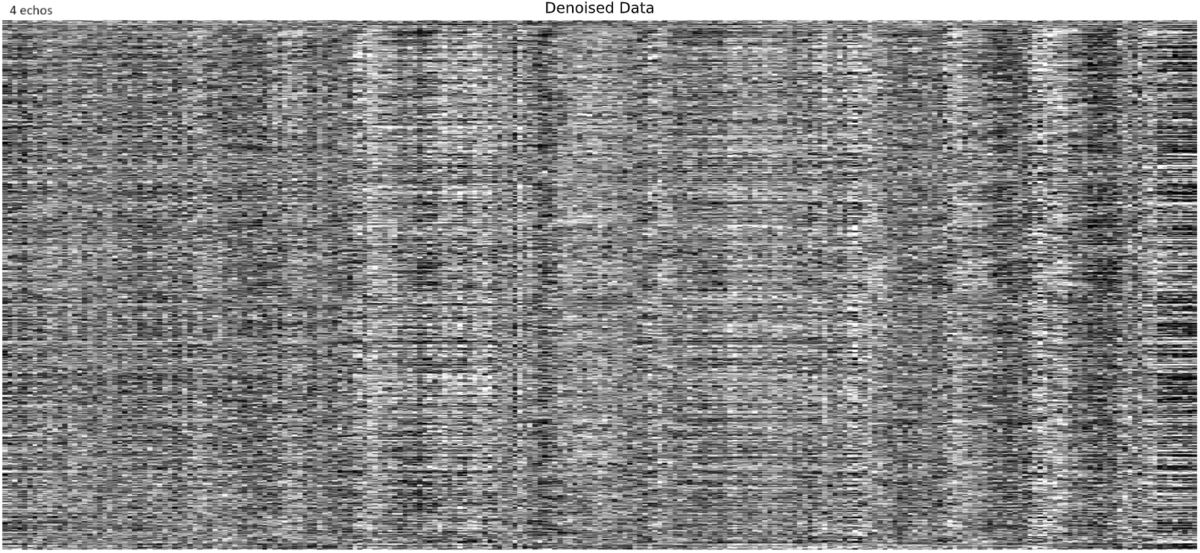
- I was worried that the high accelaration of MB4+GRAPPA2 might lead to bad motion artifacts. However, at least the carpet plots don’t show much difference compared with the MB4 protocol:
MB4 (before and after denoising)
MB4 + GRAPPA2 (before and after denoising)
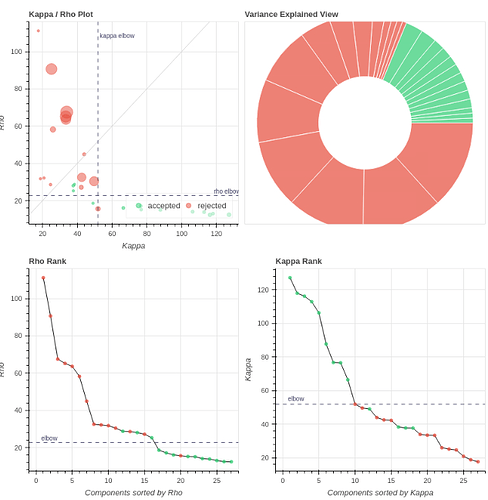
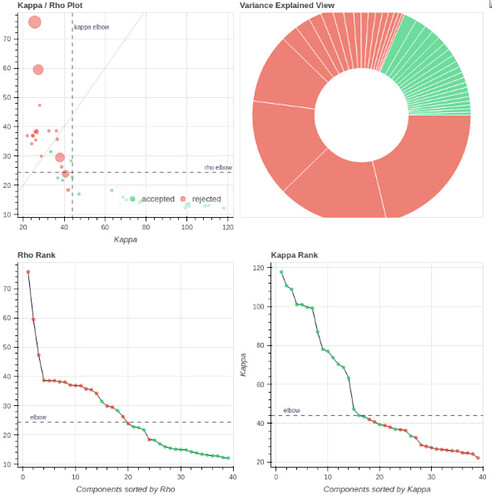
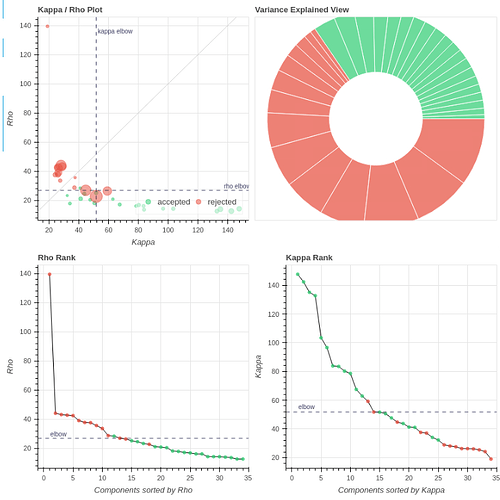
I tend towards the MB4+ GRAPPA2 protocol, since its highest echo time is lower (42 ms < 63 ms). The Adaptive Mask for both protocols is 100% at 3, meaning all voxels within the brain mask had good signal for all echos. Is this reasonable?
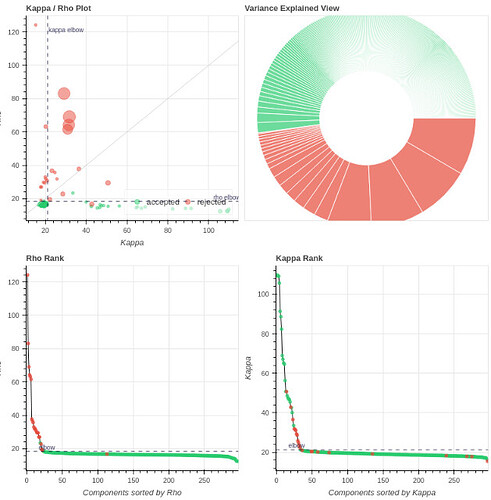
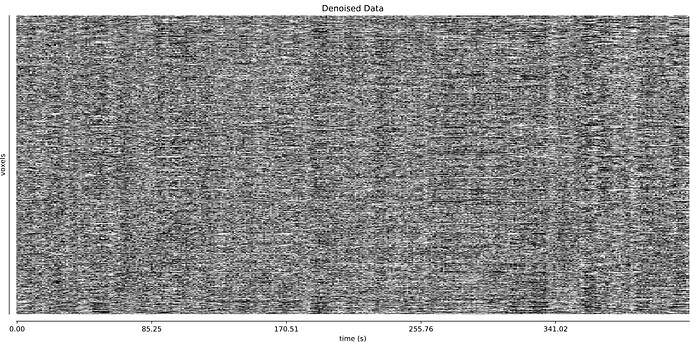
- It seems that using curve-fit has a strong impact on the denoising. Compare with the last image above:
MB4 + GRAPPA2 + curve-fit (after denoising)
Is this as surprising as I feel it is?
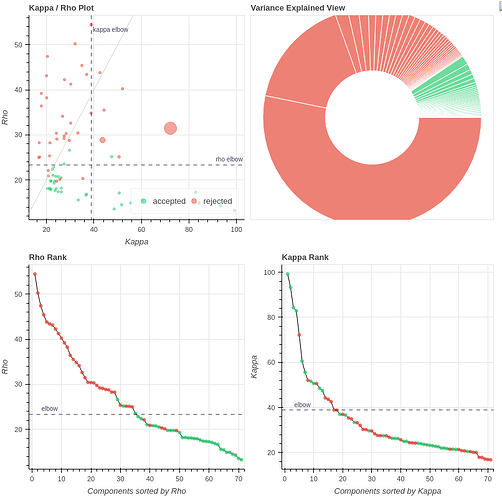
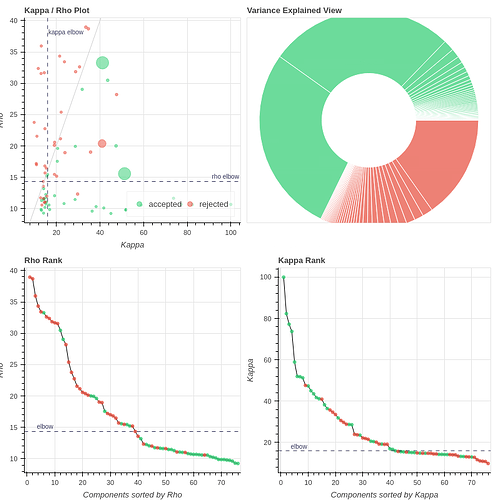
- Both protocols led to ~27 ICs, with ~20% variance explained in the final accepted ICs. Is this much lower than expected for fixation data?
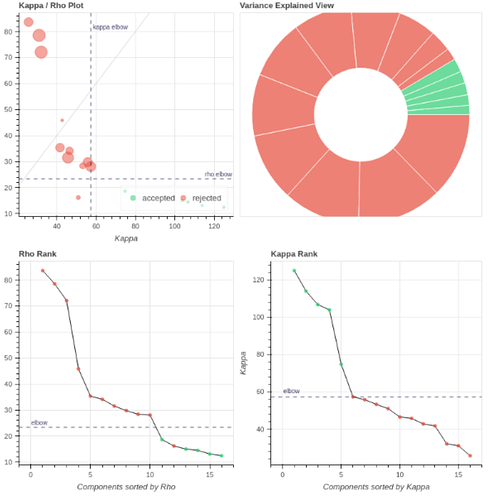
However, if I use tedpca=kundu, about 50% of the variance is retained:
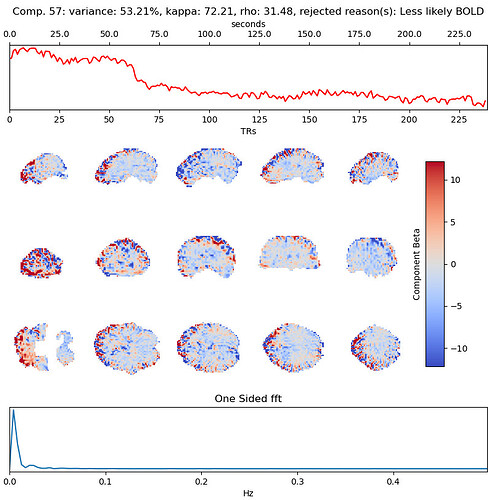
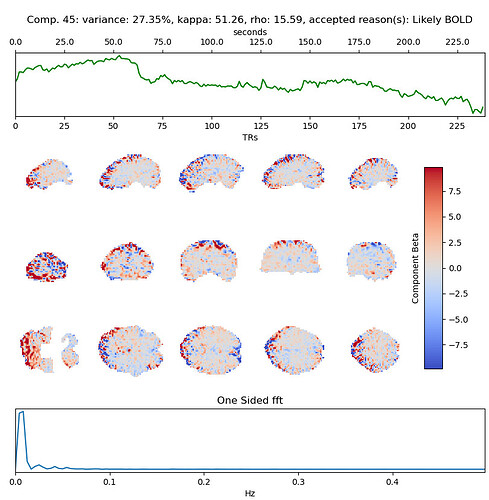
These include many ICs with a broad spectrum and very diffuse whole-brain maps, so I lean against using it here. Do you agree?
To summarize, I tend to go with MB4 + GRAPPA2 using TEDANA with curve-fit.
I would love to hear it you think I’m missing something here.
In case it’s helpful, I uploaded a Drive folder with tedana_report.html file and the figures folder for each protocol and pipeline.
Many thanks for your time!
Best,
Roey
Scan protocol
Data collected on 3T Siemens MAGNETOM Skyra scanner with 32 channel headcoils, with fixation for 7:10 minutes.
- Protocol 1 used MB accelaration factor of 4 with 3 echo times (14, 39, 63 ms). TR=1000 ms (466 volumes).
- Protocol 2 used MB accelaration factor of 4 + GRAPPA factor of 2 with 3 echo times (12, 27, 42 ms). TR=760 ms (613 volumes).