Dylan! Thanks so much for the response! I think we’re getting close, but we’re still not quite on the same page. I’ll add a code example to give you a better intuition for the problem. But first:
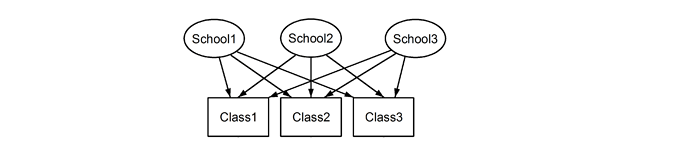
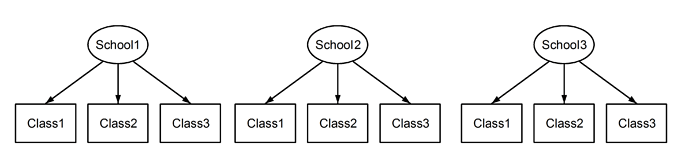
What I take that to mean is you don’t care what the mean value in each region for each transition is, you just want to see if transition has an effect on the value in each region. Is your goal to determine which regions transitions has an effect on?
Nope! It’s the opposite way around. I do care about the mean value for each region over the transitions (but I don’t care about the individual differences between transitions). Instead, you can think of the transitions as different samples for each region. Just like you would ask the question: “I know that people’s mood is different over time. But are some people happier than others across all timepoints?”
Another way to phrase it: I would like to measure the mean value for each region across transitions, to have an estimate for how ‘important’ that region is across different transitions. Since I have to assume that subjects systematically differ in their overall baselines, I (probably) need to use a method which takes that into account (aka. ask that same question for each subject individually).
Here’s a code example with a (probably dumb) method of how to do it (which does however, what I want):
import numpy as np
import pandas as pd
from scipy.stats import ttest_1samp
import seaborn as sns
# let's get some (real) example data with only 2 subjects (in reality it's ~ 500), 3 transitions,
# (in reality it's 12) and 3 regions (in reality it's 376)
df = pd.DataFrame( {'subject_id': {0: 'subject_1', 1: 'subject_1', 2: 'subject_1',
3: 'subject_1', 4: 'subject_1', 5: 'subject_1',
6: 'subject_1', 7: 'subject_1', 8: 'subject_1',
9: 'subject_2', 10: 'subject_2', 11: 'subject_2',
12: 'subject_2', 13: 'subject_2', 14: 'subject_2',
15: 'subject_2', 16: 'subject_2', 17: 'subject_2'},
'transition_name': {0: 't_1', 1: 't_1', 2: 't_1', 3: 't_2',
4: 't_2', 5: 't_2', 6: 't_3', 7: 't_3',
8: 't_3', 9: 't_1', 10: 't_1', 11: 't_1',
12: 't_2', 13: 't_2', 14: 't_2', 15: 't_3',
16: 't_3', 17: 't_3'},
'node_name': {0: 'HIP-rh', 1: 'AMY-rh', 2: 'pTHA-rh', 3: 'HIP-rh',
4: 'AMY-rh', 5: 'pTHA-rh', 6: 'HIP-rh', 7: 'AMY-rh',
8: 'pTHA-rh', 9: 'HIP-rh', 10: 'AMY-rh', 11: 'pTHA-rh',
12: 'HIP-rh', 13: 'AMY-rh', 14: 'pTHA-rh', 15: 'HIP-rh',
16: 'AMY-rh', 17: 'pTHA-rh'},
'value': {0: 18.169628442050033, 1: 2453.740158396672, 2:
23.941483293259502, 3: 65.4016090514235,
4: 3.283942478187501, 5: 67.43542817427837,
6: 2162.7234920346027, 7: 7655.829395438006,
8: 211.22679142416803, 9: 1244.4375520435424,
10: 1.4496241718039369, 11: 528.1145899402427,
12: 2077.8338111876874, 13: 550.7483033777041,
14: 185.7183214382474, 15: 861.6458143820387,
16: 159.5386394755295, 17: 67.99382988159742}} )
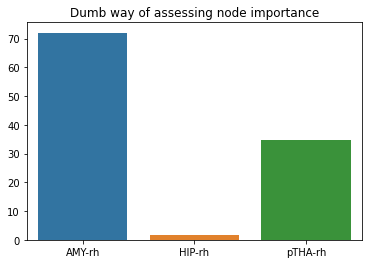
# and here's a (probably very dumb) way to do it
all_subjects_ttests = []
# iterate over each subject
for subject_id,subject_df in df.groupby('subject_id'):
# now iterate over each node within each subject and compute a one-sample
# t-test with all transitions as samples. Save the resulting t-values
single_subject_ttests = []
for node_name, node_df in subject_df.groupby('node_name'):
print(node_name)
t,_ = ttest_1samp(node_df['value'],popmean=0)
single_subject_ttests.append(t)
# save the results of each subject so that you end up with a list of lists
# of t-values
all_subjects_ttests.append(single_subject_ttests)
# now again compute t-tests using the t-values but this time take the subjects
# as samples
all_subjects_ttests = np.array(all_subjects_ttests)
ts,_ = ttest_1samp(all_subjects_ttests, popmean=0)
sns.barplot(x=['AMY-rh','HIP-rh','pTHA-rh'],y=ts).set_title('Dumb way of assessing node importance')
 I would love to use your package, so I can stick to Python and would not have to switch to R.
I would love to use your package, so I can stick to Python and would not have to switch to R.