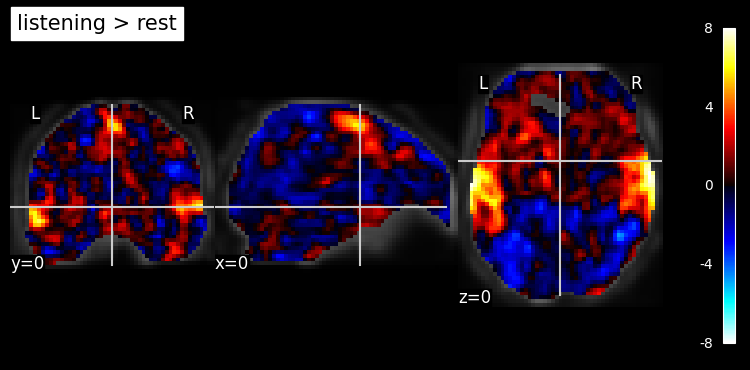
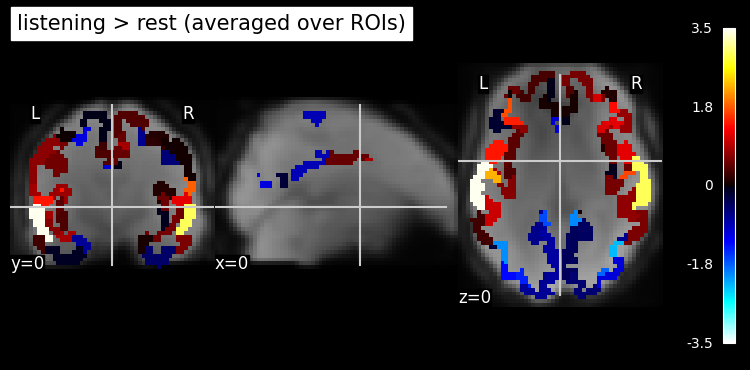
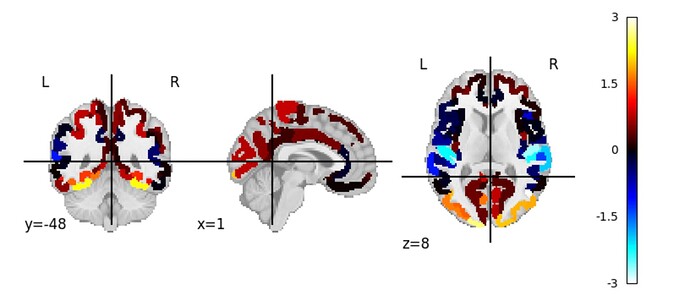
Summary of what happened:
I’m new to nilearn, so I’m struggling with finding the right documentation for doing this. I’d like to perform a typical first-level GLM analysis with a specified design matrix, but instead of fitting the model at the voxel level, I’d like to find results for the average signal in each ROI of an atlas. Ideally, I could then visualize the whole brain stat map as ROIs.
Command used (and if a helper script was used, a link to the helper script or the command generated):
So far I’ve got the atlas imported and a NiftiLabelsMasker object created with it that seems to be the right shape when I transform a nifti image with it. However, when I try to use that masker object as the ‘mask_img’ argument in a FirstLevelModel, I get an error about incompatible data. Am I misunderstanding what kind of masker is possible to pass to the GLM function, or is there something I should be doing to my data nifti first?
Thanks for your help in advance.
destrieux = nilearn.datasets.fetch_atlas_destrieux_2009()
masker = nilearn.maskers.NiftiLabelsMasker(labels_img=destrieux.maps, labels=destrieux.labels, standardize=True)
masker = masker.fit()
masker_firstlevel_glm = nilearn.glm.first_level.FirstLevelModel(mask_img=masker)
masker_firstlevel_glm = masker_firstlevel_glm.fit(smooth_brain, design_matrices=dm)
Version:
nilearn 0.11.1
Relevant log outputs (up to 20 lines):
TypeError Traceback (most recent call last)
Cell In[302], line 2
1 masker_firstlevel_glm = niglm.first_level.FirstLevelModel(mask_img=masker)
----> 2 masker_firstlevel_glm = masker_firstlevel_glm.fit(smooth_brain, design_matrices=dm)
File /opt/miniconda3/lib/python3.12/site-packages/nilearn/glm/first_level/first_level.py:884, in FirstLevelModel.fit(self, run_imgs, events, confounds, sample_masks, design_matrices, bins)
881 # Initialize masker_ to None such that attribute exists
882 self.masker_ = None
--> 884 self._prepare_mask(run_imgs[0])
886 self.design_matrices_ = self._create_all_designs(
887 run_imgs, events, confounds, design_matrices
888 )
890 # For each run fit the model and keep only the regression results.
File /opt/miniconda3/lib/python3.12/site-packages/nilearn/glm/first_level/first_level.py:1143, in FirstLevelModel._prepare_mask(self, run_img)
1128 elif not isinstance(
1129 self.mask_img, (NiftiMasker, SurfaceMasker, SurfaceImage)
1130 ):
1131 self.masker_ = NiftiMasker(
1132 mask_img=self.mask_img,
1133 smoothing_fwhm=self.smoothing_fwhm,
(...) 1141 memory_level=self.memory_level,
1142 )
-> 1143 self.masker_.fit(run_img)
1145 else:
1146 # Make sure masker has been fitted otherwise no attribute mask_img_
1147 self.mask_img._check_fitted()
File /opt/miniconda3/lib/python3.12/site-packages/nilearn/maskers/nifti_masker.py:479, in NiftiMasker.fit(self, imgs, y)
475 self.mask_img_ = self._cache(compute_mask, ignore=["verbose"])(
476 imgs, verbose=max(0, self.verbose - 1), **mask_args
477 )
478 else:
--> 479 self.mask_img_ = _utils.check_niimg_3d(self.mask_img)
481 if self.reports: # save inputs for reporting
482 self._reporting_data = {
483 "mask": self.mask_img_,
484 "dim": None,
485 "images": imgs,
486 }
File /opt/miniconda3/lib/python3.12/site-packages/nilearn/_utils/niimg_conversions.py:372, in check_niimg_3d(niimg, dtype)
338 def check_niimg_3d(niimg, dtype=None):
339 """Check that niimg is a proper 3D niimg-like object and load it.
340
341 Parameters
(...) 370
371 """
--> 372 return check_niimg(niimg, ensure_ndim=3, dtype=dtype)
File /opt/miniconda3/lib/python3.12/site-packages/nilearn/_utils/niimg_conversions.py:317, in check_niimg(niimg, ensure_ndim, atleast_4d, dtype, return_iterator, wildcards)
312 return ni.image.concat_imgs(
313 niimg, ensure_ndim=ensure_ndim, dtype=dtype
314 )
316 # Otherwise, it should be a filename or a SpatialImage, we load it
--> 317 niimg = load_niimg(niimg, dtype=dtype)
319 if ensure_ndim == 3 and len(niimg.shape) == 4 and niimg.shape[3] == 1:
320 # "squeeze" the image.
321 data = safe_get_data(niimg)
File /opt/miniconda3/lib/python3.12/site-packages/nilearn/_utils/niimg.py:126, in load_niimg(niimg, dtype)
124 niimg = load(niimg)
125 elif not isinstance(niimg, spatialimages.SpatialImage):
--> 126 raise TypeError(
127 "Data given cannot be loaded because it is"
128 " not compatible with nibabel format:\n"
129 + repr_niimgs(niimg, shorten=True)
130 )
132 dtype = _get_target_dtype(_get_data(niimg).dtype, dtype)
134 if dtype is not None:
135 # Copyheader and set dtype in header if header exists
TypeError: Data given cannot be loaded because it is not compatible with nibabel format:
destrieux2009_rois...