After reading every thread about the topic on NeuroStars, I feel more confused! ![]()
I want to run a level 1 (run level) GLM analysis in FEAT on task(event)-based multi-band BOLD data. (I have not excluded slice-timing-correction in fmriprep.)
I will share my understanding below. I wish I could include a TLDR, but this is a topic about details, so I’m afraid it will be long. I appreciate you getting through reading it at any length, and I’m looking forward to any comments/corrections on any of the mentioned points.
- I should choose “Full Analysis” rather than just “Statistics”.
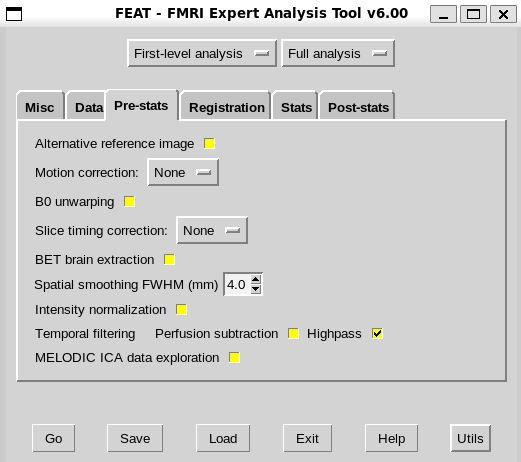
1a. Since spatial smoothing is not performed by fmriprep, I will need to apply that in the pre-stats tab.
1b. Since FSL needs registration matrices for higher levels, I can quickly register 4D data to MNI152 with 3 DOF in the registration tab of the GUI to generate these matrices (even with the workaround)
1c. Since fmriprep doesn’t apply temporal highpass filtering, I will need to apply that in the pre-stats tab unless I’m using the aCompCor+Cosine denoising strategy (as discussed and cogently explained by @paulmccarthy here)
1d. The rest of the options in Pre-stats and Registration tabs should be left inactive.
1e. Since fmriprep doesn’t apply BET of 4d data, I should include the brain mask in the .fsf file in the non-gui section at the end. FSL still won’t use the skull stripped images for the rendered_thresh_zstat visualization.
- I should turn on “FILM prewhitening” and add confound regressors as a text file as “additional confound EVs”. Then proceed with a denoising strategy that preferably covers both motion realignment and physiological signal regression (as suggested by @Steven here and compared in this chart)
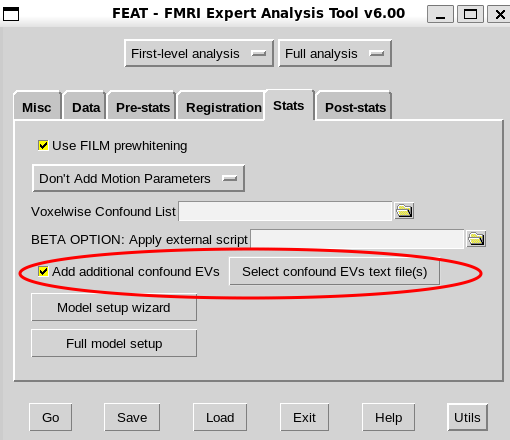
2a. for the 24p denoising strategy, the .txt file should just include the respective trans and rot columns and their expansions from the confounds.tsv file of fmriprep output. These motion regressors do not need to be detrended, demeaned, standardized, etc (not even for the powers columns?). The temporal highpass filtering option should also be applied in FEAT.
2b. for the Satterthwaite 36p denoising strategy, the .txt file should include the 24 above plus the tissue signal columns (global, wm, csf, and their expansions). The tissue signals regressors seem to need some kind of processing ( demeaned or standardized ?) since their range is vastly different than the motion params. Since GSR is still debated, maybe I can only include the csf and wm columns (and maybe their expansion). There is a csf_wm column in the confounds.tsv that I assume is the mean signal in the combined csf+wm mask, but I haven’t seen it mentioned anywhere in fmriprep documentation. I guess this could be used instead of all those signal regressor columns as a more minimal denoising strategy to spare some dof. If I include GSR, it won’t interfere with the grand-mean scaling automatically performed by FEAT. As a side note, heavy motion/tissue signal denoising is not emphasized for task-based fMRI for various reasons, including the fact that WM signal could be task related. Also ICA-based methods (e.g FIX) seem to perform quite well in identifying noise/unrelated components.
2c. for the aCompCor+Cosine denoising strategy, the .txt file should include the 24 above(though some people only include 12: the 6 motion params and their derivatives ?) plus a number of aCompCor columns (to explain a certain percentage of variance, as given in the included json files) plus all the 11 Cosine columns (though some people only apply 1 or 5 of these Cosine columns ? as described by @snastase here) . The aCompoCor and Cosine regressors do not need to be detrended, demeaned, standardized, etc (?). The motion params don’t need to be detrended either (as suggested by @pbellec here ). The temporal highpass filtering option should not be applied in FEAT due to the inclusion of Cosine regressors, but it is still fine to add temporal derivatives of original task EVs to the model.
2d. Regardless of the chosen strategy, I can safely include the one-hot non-steady-state columns in the .txt file to regress out the first few volumes of my 4D data (?)
2e. Regardless of the chosen strategy, I can safely censor/scrub outlier volumes by including the one-hot motion_outlier columns. Censoring will not interfere with the my task EVs and their temporal derivatives in the model (?)
2f. for methods that use tissue signal regression, I should also calculate and include the means of global, csf, wm columns as regressors at group level (?).