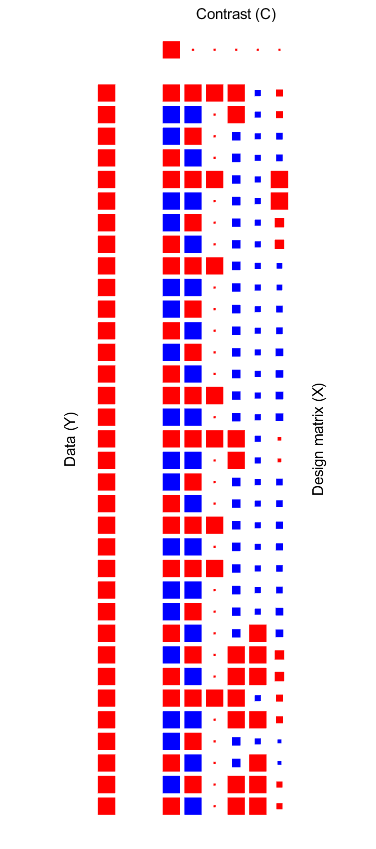
Hi everyone,
I am trying to complete resting-state connectivity analysis using CONN’s command-line GLM module (conn_module('GLM',...)) for a Treatment vs Placebo crossover design (17 subjects, 2 sessions each). My issue is that I can run the code, however, I am getting 152 roi clusters (for 4S156) which I am quite sure is incorrect. I have attached a figure of my matrix that I am passing into the and I’ll attach the code below:
C2 = [1 0 0 0 0 0]
% X2 = treatment [1=treat -1 Placebo], session [1 = ses1, -1=ses2], carry over [1 placebo second ses], then mean centered sex, recent drinking (24hrs), age
X2 = [ ...
1 1 0 0.705882353 -0.205882353 10.35294118
-1 -1 1 0.705882353 -0.205882353 10.35294118
-1 1 0 -0.294117647 -0.205882353 -9.647058824
1 -1 0 -0.294117647 -0.205882353 -9.647058824
1 1 0 -0.294117647 -0.205882353 32.35294118
-1 -1 1 -0.294117647 -0.205882353 32.35294118
-1 1 0 -0.294117647 -0.205882353 15.35294118
1 -1 0 -0.294117647 -0.205882353 15.35294118
1 1 0 -0.294117647 -0.205882353 -7.647058824
-1 -1 1 -0.294117647 -0.205882353 -7.647058824
-1 1 0 -0.294117647 -0.205882353 -9.647058824
1 -1 0 -0.294117647 -0.205882353 -9.647058824
-1 1 0 -0.294117647 -0.205882353 -11.64705882
1 -1 0 -0.294117647 -0.205882353 -11.64705882
1 1 0 -0.294117647 -0.205882353 -11.64705882
-1 -1 1 -0.294117647 -0.205882353 -11.64705882
1 1 0 0.705882353 -0.205882353 3.352941176
-1 -1 1 0.705882353 -0.205882353 3.352941176
-1 1 0 -0.294117647 -0.205882353 -9.647058824
1 -1 0 -0.294117647 -0.205882353 -9.647058824
1 1 0 -0.294117647 -0.205882353 -10.64705882
-1 -1 1 -0.294117647 -0.205882353 -10.64705882
1 1 0 -0.294117647 -0.205882353 -9.647058824
-1 -1 1 -0.294117647 -0.205882353 -9.647058824
-1 1 0 -0.294117647 -0.205882353 -11.64705882
1 -1 0 -0.294117647 0.794117647 -11.64705882
-1 1 0 0.705882353 0.794117647 15.35294118
1 -1 0 0.705882353 0.794117647 15.35294118
1 1 0 0.705882353 -0.205882353 10.35294118
-1 -1 1 0.705882353 0.794117647 10.35294118
-1 1 0 -0.294117647 -0.205882353 -3.647058824
1 -1 0 -0.294117647 0.794117647 -3.647058824
-1 1 0 0.705882353 0.794117647 8.352941176
1 -1 0 0.705882353 0.794117647 8.352941176
]
%% --- Run GLM: Treat> Placebo ---
conn_module('GLM', ...
'data', {'allsubses01ses02_4S156_connmat.nii'}, ...
'design_matrix', X2, ...
'contrast_between', C2);
Please let me know what I am doing wrong.
Thank you in advance