Hi everyone. I’d like to convert fmriprep’s *_from-T1w_to-fsnative_mode-image_xfm.txt file to *.h5 format so that I can apply the transformation using the Python implementation of ANTs’ function apply_to_image(). I’ve been trying to use the h5py package in Python to accomplish this, but I haven’t been able to get it working. Any suggestions? See below for details.
It fails at sub_to_fsnative = ants.read_transform(trans_h5) with the following error:
"Traceback (most recent call last):
File "<stdin>", line 1, in <module>
File "/Users/flutist4129/Library/Python/3.9/lib/python/site-packages/ants/core/ants_transform_io.py", line 330, in read_transform
dimensionUse = libfn1(filename)
RuntimeError: /Users/admin/actions-runner/_work/ANTsPy/ANTsPy/itksource/Modules/IO/TransformBase/src/itkTransformFileReader.cxx:128:
ITK ERROR: TransformFileReaderTemplate(0x60004e7a2600): Could not create Transform IO object for reading file /Users/flutist4129/Documents/Northwestern/studies/mwmh/data/processed/neuroimaging/surf/sub-MWMH212/ses-2/anat/sub-MWMH212_ses-2_from-T1w_to-fsnative_mode-image_xfm.h5
Tried to create one of the following:
HDF5TransformIOTemplate
HDF5TransformIOTemplate
MatlabTransformIOTemplate
MatlabTransformIOTemplate
TxtTransformIOTemplate
TxtTransformIOTemplate
You probably failed to set a file suffix, or
set the suffix to an unsupported type."
import os
import json
import sys, getopt
import argparse
import ants
import numpy as np
import h5py
parser = argparse.ArgumentParser()
parser.add_argument('-i')
parser.add_argument('-o')
parser.add_argument('-s')
parser.add_argument('-ss')
parser.add_argument('-t', nargs='+')
args = parser.parse_args()
indir = args.i
outdir = args.o
sub = args.s
ses = args.ss
tasks = args.t
# Directory where preprocessed fMRI data is located
subindir = os.path.join(indir, sub)
sesindir = os.path.join(subindir, ses)
funcindir = os.path.join(sesindir, 'func')
# Directory where outputs should go
suboutdir = os.path.join(outdir, sub)
sesoutdir = os.path.join(suboutdir, ses)
os.makedirs(os.path.join(outdir, sub, ses), exist_ok=True)
# Get the number of sessions
numses = 0
for root, dirs, files in os.walk(subindir):
for dir in dirs:
if dir.startswith('ses'):
numses = numses + 1
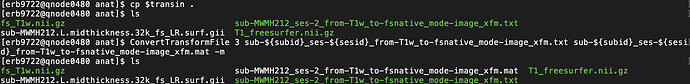
# Load Freesurfer's T1w image (fsnative)
if numses == 1:
fs_T1 = ants.image_read(f"{sesoutdir}/anat/fs_T1w.nii.gz")
elif numses == 2:
fs_T1 = ants.image_read(f"{suboutdir}/anat/fs_T1w.nii.gz")
else:
raise ValueError("There is some number of sessions other than 1 or 2.")
# Set up file paths to transformation txts
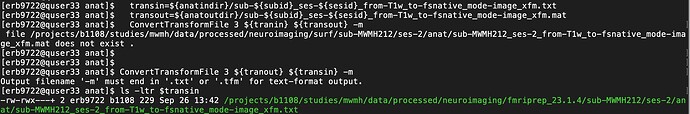
trans_txt = f"{sub}_{ses}_from-T1w_to-fsnative_mode-image_xfm.txt" #sub-{sub}_from-T1w_to-MNI152NLin6Asym_mode-image_xfm.h5
trans_h5 = f"{sub}_{ses}_from-T1w_to-fsnative_mode-image_xfm.h5"
if numses == 1:
trans_txt = f"{sesindir}/anat/{trans_txt}"
trans_h5 = f"{sesoutdir}/anat/{trans_h5}"
elif numses == 2:
trans_txt = f"{subindir}/anat/{trans_txt}"
trans_h5 = f"{suboutdir}/anat/{trans_h5}"
else:
raise ValueError("There is some number of sessions other than 1 or 2.")
# Process the file to extract the transformation parameters
parameters = []
with open(fp_txt, 'r') as file:
for line in file:
if line.startswith('Parameters:'):
# Extract numbers from this line
numbers = line.split()[1:] # Skip the "Parameters:" part
parameters = [float(num) for num in numbers]
break # Assuming only one set of parameters is needed
# Convert the list of parameters into a NumPy array
sub_to_fsnative = np.array(parameters).reshape(3, 4) # Reshape according to your matrix shape, here it's assumed to be 3x4
# Create an HDF5 file and write the matrix to it
with h5py.File(trans_h5, 'w') as hdf:
hdf.create_dataset('sub_to_fsnative', data=sub_to_fsnative)
sub_to_fsnative = ants.read_transform(trans_h5)
for task in tasks:
# Load input image
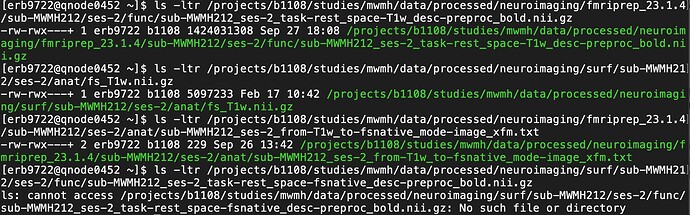
input_path = f"{funcindir}/{sub}_{ses}_{task}_space-T1w_desc-preproc_bold.nii.gz"
input = ants.image_read(input_path)
# Apply transform and write out transformed image
output = sub_to_fsnative.apply_to_image(input, reference=fs_T1)
output_path = f"{sesindir}/func/{sub}_{ses}_{task}_space-fsnative_desc-preproc_bold.nii.gz"
ants.image_write(output, output_path)