Hi, @Relyativist–
Hmm, interesting, OK.
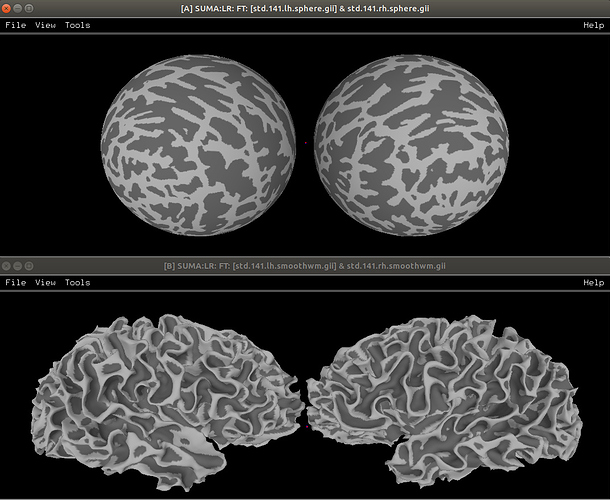
AFNI’s @SUMA_Make_Spec_FS translates a lot of FreeSurfer’s recon-all into standard format files; for example, surfaces can (should?) be converted to NIFTIs. This includes a sphere representation of each hemisphere, such as in the top panel here, which are topologically the same as the more anatomically correct lower panel surface hemispheres (viewed in SUMA):
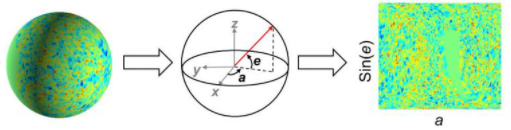
I guess the Mercator projection is just a formula for mapping points p_{\rm sph} = (\phi, \theta) on a unit sphere to a point on a rectangular grid p_{\rm rect}= (x, y). I guess I would approach this by dumping the points in the sphere (above), recentering them and normalizing them to a unit sphere; that gives you p_{{\rm sph}, i} for each $i$th node, and you could map that to p_{{\rm rect},i}. After that, you could map any node on that standard mesh to a point in your rectangular coordinates.
This command is an example of dumping each each node the coarser of the default standard meshes that @SUMA_Make_Spec_FS generates to mainly a text file (with some header info) that has 4 columns (node_index X Y Z):
SurfaceMetrics -coords -i_gii std.60.lh.sphere.gii -prefix std.60.lh.sphere
… which are stored in a new file called “std.60.lh.sphere.coord.1D.dset”, that can be dumped, without commented header info, into a new text file via, say:
1dcat std.60.lh.sphere.coord.coord.1D.dset > std.60.lh.sphere.coord.coord.dat
I was hoping I could find an existing program to scale this sphere, while keeping the same mesh, so the dumped points could be a unit sphere centered around the origin, but I haven’t found how to do that yet… at the moment, you could find the center of mass of the dumped coordinates, which will be a good estimate for the center, and shift that to the origin; then divide by the average distance to that new origin (which will be a good estimate of the radius). Then you would have your nodes all on a unit sphere, and be read to project away, while being able to project any other information at those same nodes in other corresponding meshes, too.
–pt