Hi,
Been testing tedana lately, using the preprocessed echos produced by fmriprep using the --me-output-echos option.
When projecting the optimally combined data resulting from tedanas pipeline to T1w-space (using the affine “scanner-to-T1w”-transforms supplied by fMRIprep), I noted some slight but systematic differences between tedana’s output and the optimally combined T1w-space data produced within the fMRIprep-workflow (i.e. *_space-T1w_desc-preproc_bold.nii.gz). Initially I thought this could be due to several factors (tedana uses a different mask etc.), but after some simple sanity checking I can now recreate the issue using data from the fMRIprep workflow only.
Here’s a distilled version of the setup:
-
fMRIprep (21.0.1) with
--output-spaces T1w func. SDC using pepolar approach (BOLD EPI PE-direction = AP) -
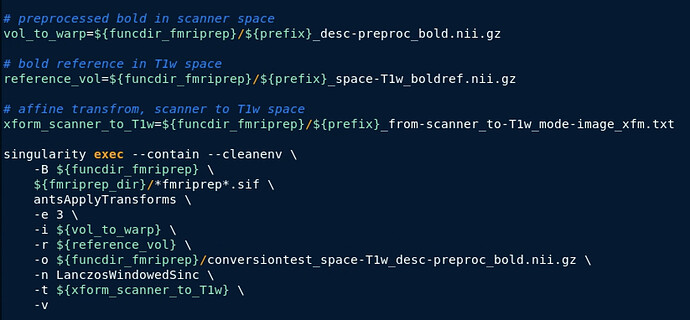
antsApplyTransform to bring preprocessed data in func/scanner space to T1w space (similar to @tsalo’s recent PR https://github.com/ME-ICA/tedana/pull/847) :
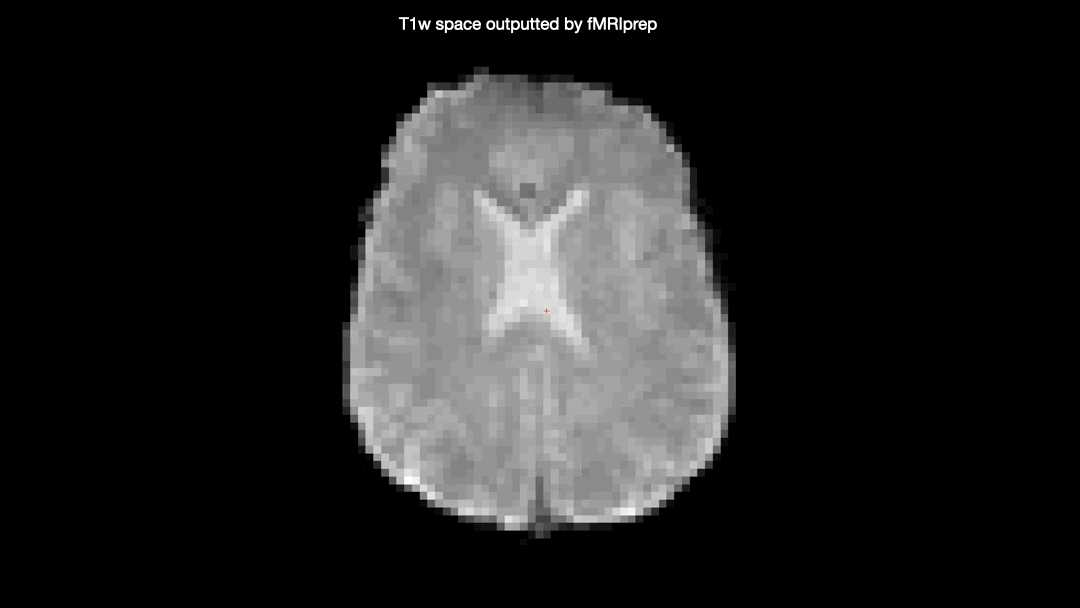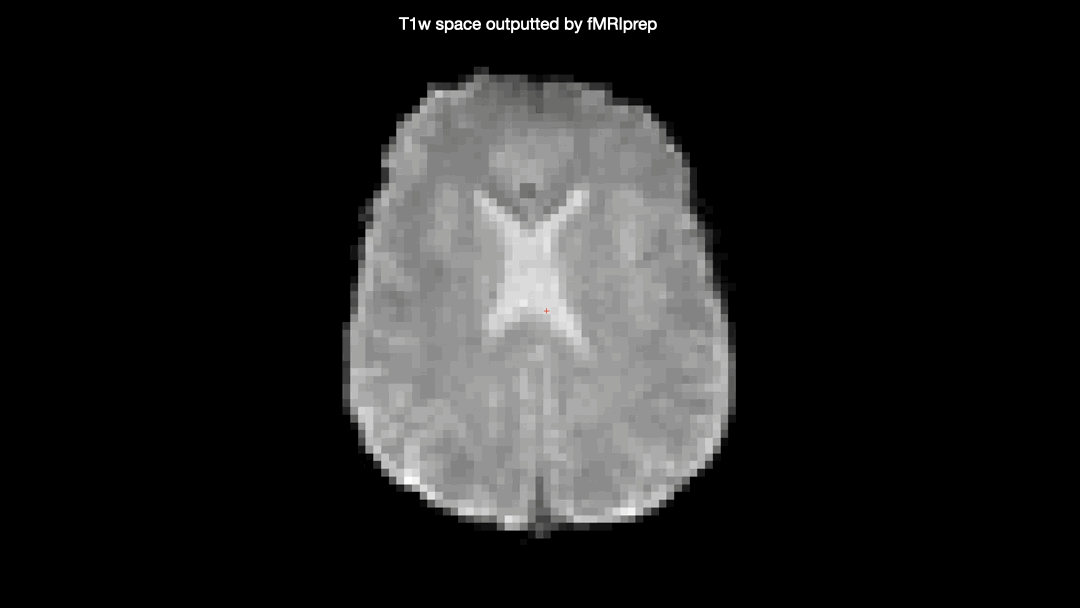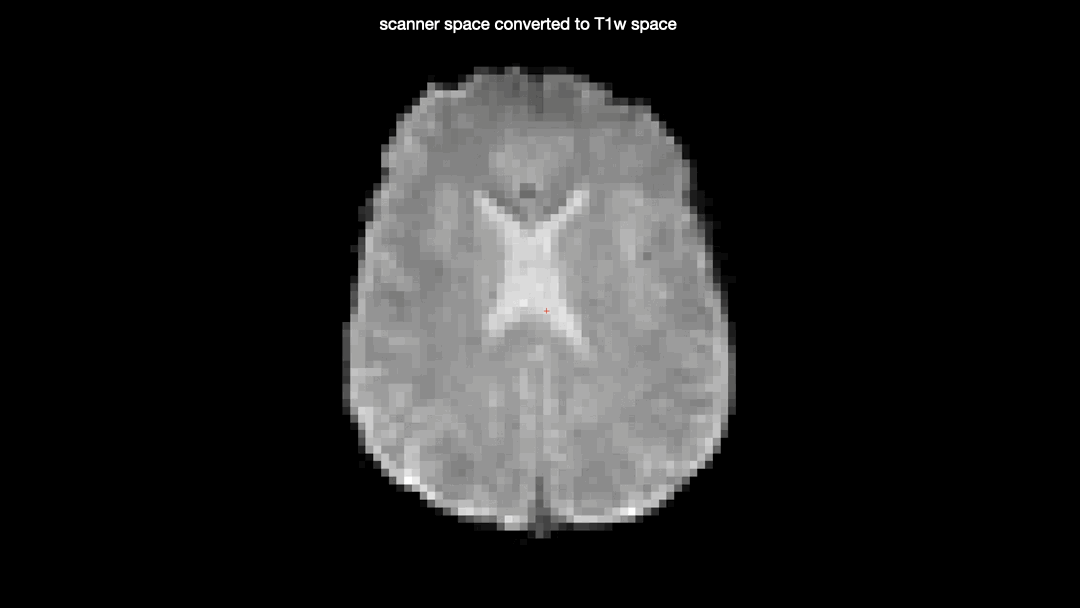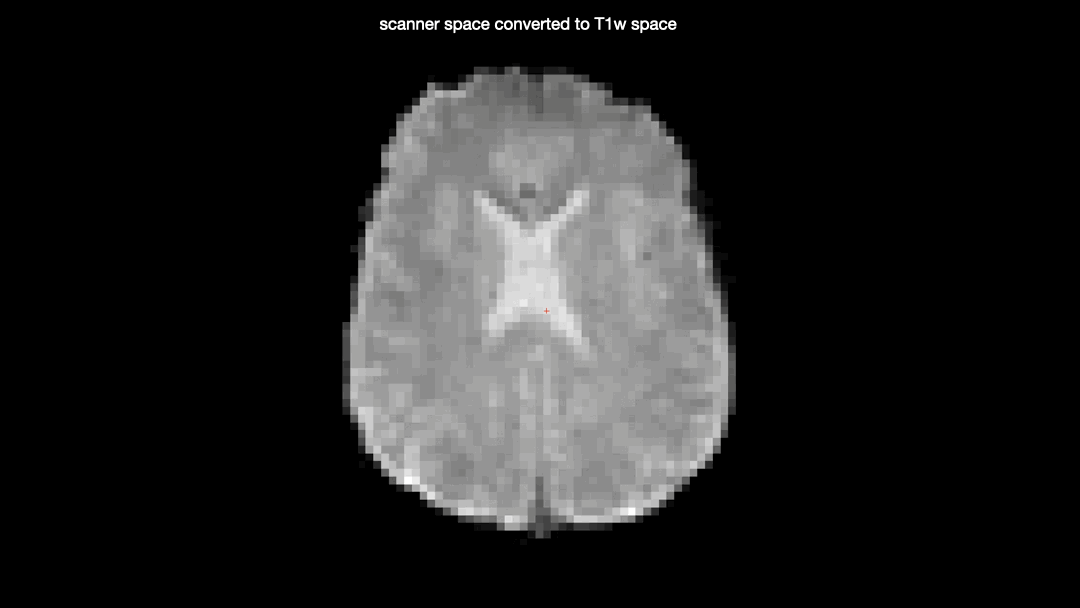
Here’s a gif comparing the converted data (scanner space → T1w space) with the T1w space data produced by fMRIprep
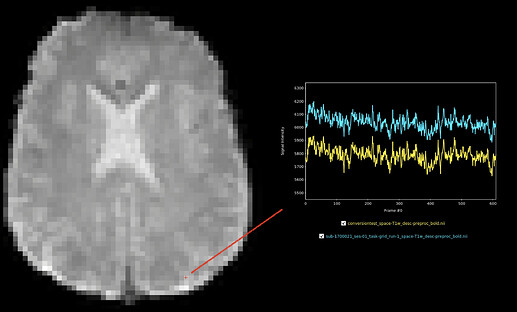
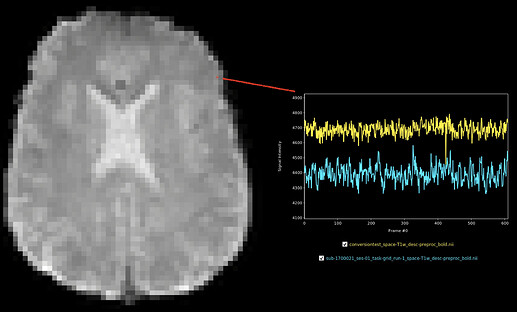
One would not expect the two datasets to be identical due to the additional interpolation-step taken on the converted data, but there seems to be some nonlinear differences as well, producing almost full “voxel shifts” anteriorly but only minor differences posteriorly.
Posterior voxel: scale shift, but highly correlated timeseries:
Anterior voxel (within cortical ribbon). Uncorrelated timeseries.
The non-linear looking differences between the two volumes almost makes me wonder if there could be differences in the SDC-steps taken for the --output-spaces T1w and the --output-spaces func workflows. However, before going into full debug-mode I wanted to check with the users/developers if I’m missing or misunderstanding something obvious here.
Thanks!