Summary of what happened:
Hi everyone
I encountered an issue while using xcp_d, and the error message is printed in the terminal window:
The above exception was the direct cause of the following exception:
Traceback (most recent call last):
File "/usr/local/miniconda/lib/python3.8/site-packages/xcp_d/cli/run.py", line 578, in main
xcpd_wf.run(**plugin_settings)
File "/usr/local/miniconda/lib/python3.8/site-packages/nipype/pipeline/engine/workflows.py", line 638, in run
runner.run(execgraph, updatehash=updatehash, config=self.config)
File "/usr/local/miniconda/lib/python3.8/site-packages/nipype/pipeline/plugins/base.py", line 224, in run
raise error from cause
RuntimeError: 44 raised. Re-raising first.
During handling of the above exception, another exception occurred:
Traceback (most recent call last):
File "/usr/local/miniconda/bin/xcp_d", line 8, in <module>
sys.exit(main())
File "/usr/local/miniconda/lib/python3.8/site-packages/xcp_d/cli/run.py", line 664, in main
failed_reports = generate_reports(
File "/usr/local/miniconda/lib/python3.8/site-packages/xcp_d/interfaces/report_core.py", line 157, in generate_reports
exsumm.collect_inputs()
File "/usr/local/miniconda/lib/python3.8/site-packages/xcp_d/interfaces/execsummary.py", line 195, in collect_inputs
task_entity_sets = task_entity_sets.sort_values(by=task_entity_sets.columns.tolist())
File "/usr/local/miniconda/lib/python3.8/site-packages/pandas/core/frame.py", line 5454, in sort_values
by = by[0]
IndexError: list index out of range
Sentry is attempting to send 50 pending error messages
Command used (and if a helper script was used, a link to the helper script or the command generated):
docker run --rm -v E:/GL/temp:/wd -v E:/GL/data135_out_fmriprep:/data -v E:/GL/xcpd_out_globalsignal_data135:/out -v D:/fantest/:/freesurfer_license pennlinc/xcp_d:0.6.0 /data /out participant -w /wd -p aroma --fd-thresh 0
--despike --lower-bpf 0.01 --upper-bpf 0.1 --smoothing 6 --cifti
Version:
xcpd:0.6.0
Environment (Docker, Singularity / Apptainer, custom installation):
Docker
Data formatted according to a validatable standard? Please provide the output of the validator:
PASTE VALIDATOR OUTPUT HERE
Relevant log outputs (up to 20 lines):
The output log for an individual participant:
Node: xcpd_wf.single_subject_MDD119_wf.cifti_postprocess_1_wf.prepare_confounds_wf.generate_confounds
Working directory: /wd/xcpd_wf/single_subject_MDD119_wf/cifti_postprocess_1_wf/prepare_confounds_wf/generate_confounds
Node inputs:
TR = 2.0
band_stop_max = None
band_stop_min = None
custom_confounds_file = None
fd_thresh = 0.0
fmriprep_confounds_file = /data/sub-MDD119/func/sub-MDD119_task-rest_acq-pa_run-1_desc-confounds_timeseries.tsv
fmriprep_confounds_json = /data/sub-MDD119/func/sub-MDD119_task-rest_acq-pa_run-1_desc-confounds_timeseries.json
head_radius = 50.0
in_file = /data/sub-MDD119/func/sub-MDD119_task-rest_acq-pa_run-1_space-fsLR_den-91k_bold.dtseries.nii
motion_filter_order = 4
motion_filter_type = None
params = aroma
Traceback (most recent call last):
File "/usr/local/miniconda/lib/python3.8/site-packages/nipype/pipeline/plugins/multiproc.py", line 67, in run_node
result["result"] = node.run(updatehash=updatehash)
File "/usr/local/miniconda/lib/python3.8/site-packages/nipype/pipeline/engine/nodes.py", line 527, in run
result = self._run_interface(execute=True)
File "/usr/local/miniconda/lib/python3.8/site-packages/nipype/pipeline/engine/nodes.py", line 645, in _run_interface
return self._run_command(execute)
File "/usr/local/miniconda/lib/python3.8/site-packages/nipype/pipeline/engine/nodes.py", line 771, in _run_command
raise NodeExecutionError(msg)
nipype.pipeline.engine.nodes.NodeExecutionError: Exception raised while executing Node generate_confounds.
Traceback:
Traceback (most recent call last):
File "/usr/local/miniconda/lib/python3.8/site-packages/nipype/interfaces/base/core.py", line 397, in run
runtime = self._run_interface(runtime)
File "/usr/local/miniconda/lib/python3.8/site-packages/xcp_d/interfaces/censoring.py", line 501, in _run_interface
confounds_df, confounds_metadata = load_confound_matrix(
File "/usr/local/miniconda/lib/python3.8/site-packages/xcp_d/utils/confounds.py", line 458, in load_confound_matrix
ica_mixing_matrix = _get_mixing_matrix(img_file)
File "/usr/local/miniconda/lib/python3.8/site-packages/xcp_d/utils/confounds.py", line 499, in _get_mixing_matrix
raise FileNotFoundError(f"Could not find mixing matrix for {img_file}")
FileNotFoundError: Could not find mixing matrix for /data/sub-MDD119/func/sub-MDD119_task-rest_acq-pa_run-1_space-fsLR_den-91k_bold.dtseries.nii
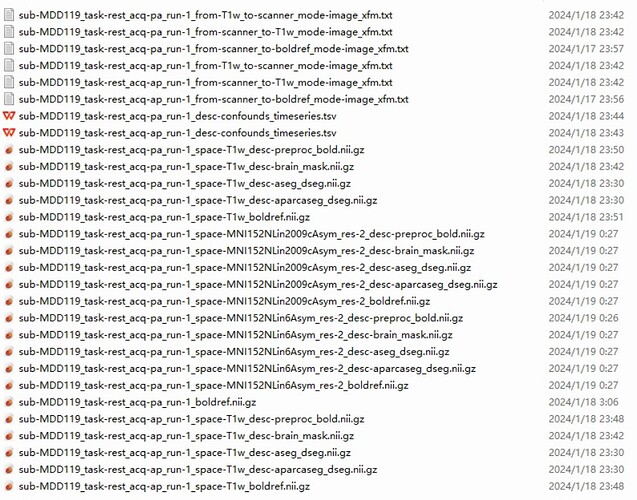
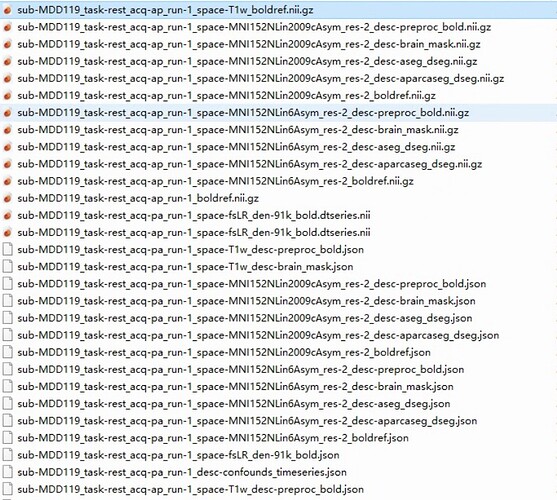
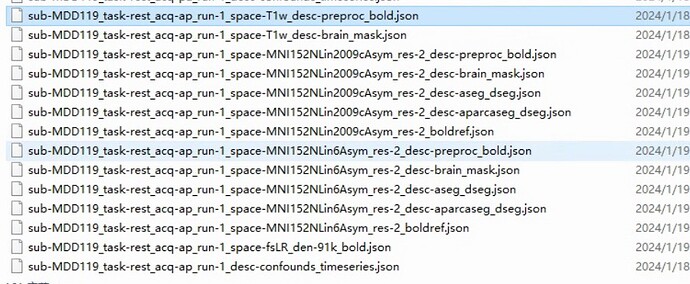
Screenshots / relevant information:
As shown in the figure, the fmriprep output results file for one of the participants.