I tried creating a mask using the Harvard-oxford cortical atlas in fsl but I realized all the the structures are for both hemispheres.
I want to know if there is a way to restrict the mask to one hemisphere. I tried using fslmaths but was not successful
Hi @Reubebe,
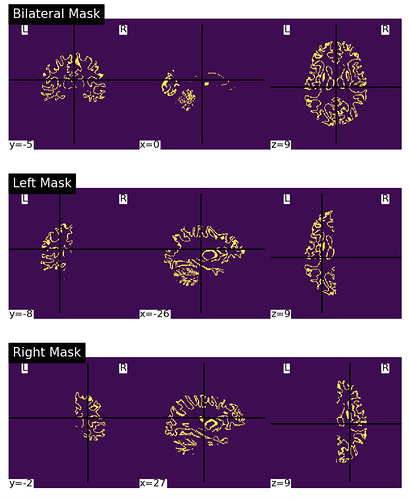
You can load your bilateral masks in Nilearn, and do something such as setting all values on the first or second half of x values to 0, and then save out the modified images as left and right masks.
There may be a more straightforward way using fslmath especially if a hemispheric mask is available, and my method assumes that the brain is centered (which is a safe assumption for most atlases), but it should work.
Best,
Steven
Here is code example
import nilearn.image
import nilearn.plotting
import matplotlib.pyplot as plt
# Load Image
bilateral_mask_path = '/path/to/mask.nii.gz'
bilateral_mask = nilearn.image.load_img(bilateral_mask_path)
mask_data = bilateral_mask.get_fdata()
affine = bilateral_mask.affine
# Get center X-coord
x_dim = mask_data.shape[0]
x_center = int(x_dim/2)
nilearn.plotting.plot_img(bilateral_mask, title='Bilateral Mask')
plt.show()
# Get left mask
mask_data_left = mask_data.copy()
mask_data_left[0:x_center,:,:] = 0
mask_left = nilearn.image.new_img_like(bilateral_mask, mask_data_left, affine=affine, copy_header=True)
nilearn.plotting.plot_img(mask_left, title='Left Mask')
plt.show()
mask_left.to_filename('left_mask.nii.gz')
# Get right mask
mask_data_right = mask_data.copy()
mask_data_right[x_center:,:,:] = 0
mask_right = nilearn.image.new_img_like(bilateral_mask, mask_data_right, affine=affine, copy_header=True)
nilearn.plotting.plot_img(mask_right, title='Right Mask')
plt.show()
mask_right.to_filename('mask_right.nii.gz')
2 Likes