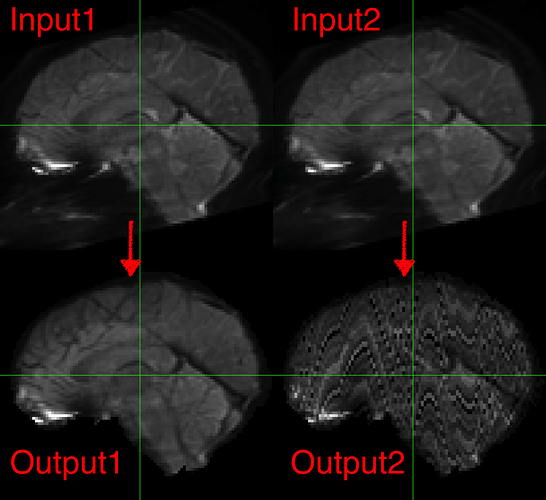
I am trying to use csdsi_3dshore recon to reconstruct DSI data into HCP-like shelled data to use with MRtrix, but the output is “scrambled” (“Output2” bottom right). I know this pipeline is listed as experimental, so I might have just given up, but one time when I ran it, it seemed to have worked? (“Output1” bottom left)
I was messing with so many things trying to debug various aspects of this dataset that I can’t tell what magically caused it to work that time but never since. The preproc outputs in output/qsiprep/, which are supposedly the inputs to the “recon_shore” node do not look identical, but neither one is obviously corrupted or anything like that (Input1 and Input2). The other input options listed for that node in its work directory are identical for both runs.
Does anybody here have any insight into what combination of options or preprocessing might cause csdsi_shore to fail in this way? Or maybe provide some guidance for how I could find the difference based on my output/work directories?
Both of the outputs shown were created using qsiprep 0.20.0. I’m running 0.21.3 and 0.21.4 now to check, though it required some patching, so we’ll see if it crashes.