I don’t see CT in the anatomical imaging section of BIDS. This is for iEEG where the CT contains the information about the placement of electrodes. Is this supported yet?
It looks like this is a BIDS Extension Proposal for CT under development here. Unfortunately, it appears to be in very early stages, so, while you can definitely contribute to it, I don’t think it’s ready for use.
I don’t know much about iEEG, but would you be able to use the coordsystem file (combined with a best-guess approximation of BIDS convention for the associated CT nifti file) for your purposes? By best-guess, I would say you could put the CT file in a new ct/ folder (as in BEP024) with a _ct suffix and all of the typical associated metadata in a json. You’d need to add CT files to your .bidsignore, though, if you’re planning to share the data or validate the dataset.
dcm2niix has json files that look like this
{
“Modality”: “CT”,
“Manufacturer”: “Philips”,
“ManufacturersModelName”: “Ingenuity_CT”,
“DeviceSerialNumber”: “300003”,
“BodyPartExamined”: “BRAIN”,
“PatientPosition”: “HFS”,
“SoftwareVersions”: “4.0”,
“SeriesDescription”: “STEREO_HEAD_iQ2_iDose_(2)”,
“ProtocolName”: “STEREO_HEAD_iQ2_iDose_(2)”,
“ScanOptions”: “HELIX”,
“ImageType”: [“ORIGINAL”, “PRIMARY”, “AXIAL”],
“SeriesNumber”: 201,
“AcquisitionTime”: “15:17:58.000000”,
“AcquisitionNumber”: 1,
“XRayExposure”: 200,
“ReconMatrixPE”: 512,
“ImageOrientationPatientDICOM”: [
1,
0,
0,
0,
1,
0 ],
“ConversionSoftware”: “dcm2niix”,
“ConversionSoftwareVersion”: “v1.0.20190902”
}
so I assume Chris Rorden has thought about it and implemented a somewhat BIDS compatible solution and it looks like this is about what will be required when the extension is released.
Just leaving this here in case it’s helpful to anyone else querying the same things.
alexrockhill as a starting point dcm2niix converts some of the DICOM tags from the CT Image Module to the BIDS sidecar. BIDS uses a NIfTI format image with a JSON format sidecar. The NIfTI format was developed by experts in MRI. The two challenges with using tools developed for MRI for the CT modality is that CT scans can often have a gantry tilt applied and CT scans often have variable slice thickness (with more slices near the survival critical brain stem and fewer in the superior cortex).
- Gantry tilt is a shear across slices, as shown in the figure below. In theory we can store this in the NIfTI S-Form, but the NIfTI Q-Form requires rectangular volumes. Further, many MRI tools, including all ITK tools assume rectangular volumes (this explains why tools like ANTS give precedence to the Q-Form).
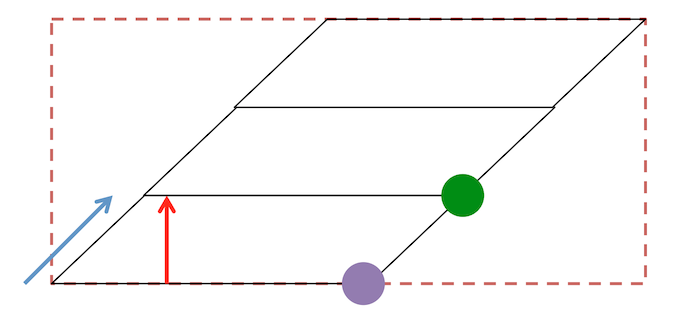
- The NIfTI format requires the the distance between voxel centers is consistent for each axis of the volume. Therefore, it is unable to handle the situation where slices are thinner in the brain stem relative to more superior slices.
dcm2niix resolves these two situations by interpolating the DICOM data into a rectangular, regularly spaced grid. This is a required deviation from dcm2niix’s typical lossless conversion. While this allows users to reliably use common MRI tools for this modality, we need to be aware of the cost for this interpolation. If one wants a BIDS specification for the CT modality, is the NIfTI format the correct one to use, when it is inherently unable to losslessly store properties of the CT modality? It is an expedient choice, but I think real consideration needs to be given to the long term implications. Consider a repository like OpenNeuro where users can store, share and replicate datasets. Given the expense of running participants (in terms of time, cost of acquistion, curation, etc.) do we really want to store interpolated data in our repositories?
In most stereo EEG/ECoG electrode localization methods papers I’ve seen (e.g. https://www.frontiersin.org/articles/10.3389/fninf.2017.00062/full) the DICOMs are converted to nii and the localizations are carried out from there. I’m not sure how much of a loss in accuracy there is due to that, I’m not sure the comparison between conversion to nii and not has been made. @Chris_Rorden would you suggest storing them in another format (I assume DICOM is too unstandardized to be adopted by BIDS likely maybe)? In the BIDS proposal to add CTs, currently it looks like the idea is to store them in nii https://docs.google.com/document/d/1fqnJZ18x5LJC8jiJ8yvPHUGFzNBZ6gW2kywYrUKWtuo/edit#
My sense is there needs to be discussion about how to handle these cases.
- One option is to demand that images are acquired without gantry tilt and with equidistant slice spacing, but this may preclude many clinically relevant datasets. Benefit: compatible with existing tools, easy to define in a specification. Cost: not all acquired data will be suitable for the archives. Image acquisition will be constrained by the BIDS standard.
- A second option would be to consider an image format other than NIfTI that can support these features. Benefit: can choose format that intrinsically supports CT features. Cost: requires all tools in pipeline support features.
- A third option would be to carefully document how to handle these situations. In other words, if the gantry tilt is not zero, the qform_code should be set to NIFTI_XFORM_UNKNOWN, and all tools used in CT pipelines must be able to at least detect shear in the S_Form. Likewise, variable slice spacing could be saved BIDS sidecar. Presumably pixdim[3] (slice thickness) in the header is set to zero to encourage the software to look in the sidecar. There seems to be a precedence for this, as the PET proposal BEP009 suggests using the tag
FrameTimesStartin the BIDS sidecar to deal with irregular sampling in the 4th dimension (time, pixdim[4]). Presumably, all tools need to inherit the value. Benefit: easy to define as a standard, lossless storage. Cost: requires all tools in pipeline support features. - The final option would be to demand that images acquired with gantry tilt or uneven slice spacing be interpolated prior to being saved as BIDS images. A core question is what interpolation should be used (dcm2niix uses linear interpolation which can lose high frequency but avoids ringing), and how does one select the resampled output slice thickness (dcm2niix resamples to the mean slice thickness, but one could argue for creating larger files that resample to the minimum slice thickness). Benefit: works well with existing tools. Cost: interpolation choices irretrievably modifies stored images.
I know a lot of CT users are happy simply using the interpolated output from dcm2niix because it just works. However, I made arbitrary decisions regarding how to handle these situations that have clear implications. If everyone agrees these choices reflect a good compromise, we can proceed with option 4. However, I think the community needs to carefully consider these options and the respective consequences of each before choosing the foundation for future data repositories that will reflect a huge investment.
Maybe there ought to be a meeting to discuss this.
I think there are good options, for the purposes I use CTs for; localizing intracranial EEG, it would be ideal to be able to visualize the shear form with unaltered slice density at least until the dcm2niix version or others that just work are shown to just work the same after interpolation within the margin of error we care about.