I am learning how to apply nilearn.signal.clean to my dataset. From my understanding, the function uses cubic spline interpolation as the only form of interpolation possible. However, if you remove something like 30 contiguous seconds from the middle of an fMRI time series, the cubic spline has really strong excursions from zero. This makes for some weird looking datasets, which after the full cleaning run such as filtering and confound regression, seems to shift the temporal position of some of the signal peaks and also smooth others, when compared to no censoring/interpolation done. As less volumes are censored, the cubic interpolated cleaned time series seems to get closer and closer to the same form of the non-censored cleaned time series. From briefly reading a few papers, it seems that choice of interpolation method seems to depend on the ultimate effect on the dataset, and the cubic spline has some advantages in regards to “Runge’s Phenomenon”.
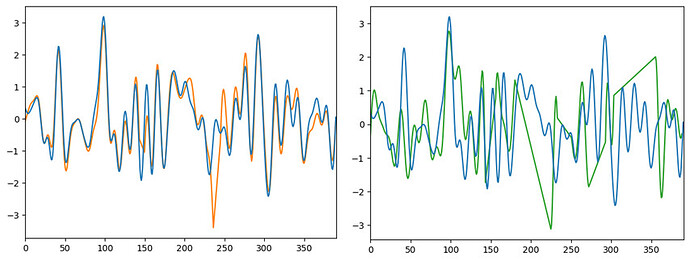
Here I’ve recreated two outputs of the same vertex using nilearn.signal.clean. It’s not very clear, but the first figure has two blocks of 10-seconds censored, and the second has a few longer 30-second+ censored (the linear portions are the censored portions). The orange figure clearly looks very similar to the non-censored data in the non-censored portions, and very different in the censored portions. The green figure looks maybe 50% different than the noncensored data. I hope I’ve written my thoughts out clearly.
I’m curious as to why cubic spline interpolation was chosen as the only option, as opposed to say, allowing for option to choose linear interpolation or some other polynomial interpolation and choosing whichever best suits the dataset, which are in the same scipy or numpy package that CubicSpline is pulled from.