Hi Neurostars Experts,
Our lab has been using dcm2bids to organize data into BIDS format, and it’s been working great for us. However, we recently encountered an issue with fMRI data collected in a new project. The data was provided in the “dicom_classic” format, where each slice is stored as a separate file, resulting in 19,836 files for a single fMRI run.
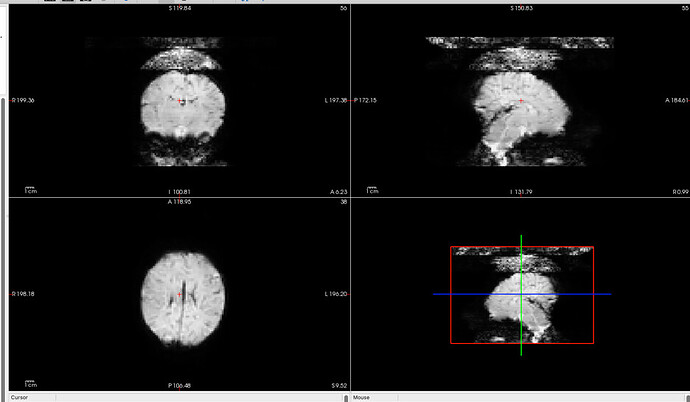
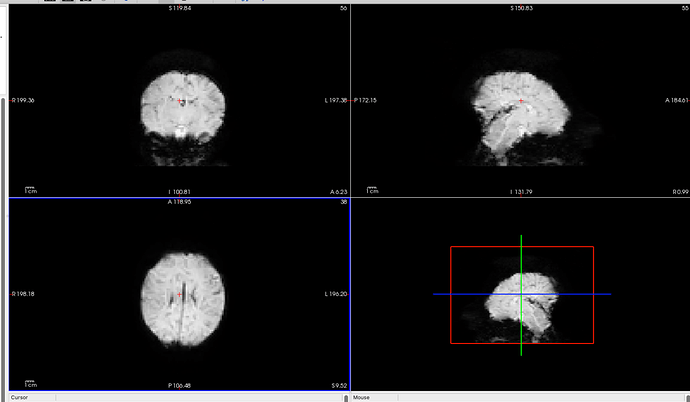
When we use dcm2bids to convert and organize this data, we notice odd artifacts on the top of the head in the fMRI images. Interestingly, when we use dcm2niix directly to convert the files, the artifacts are not present. This issue only occurs with data in “dicom_classic” format and does not happen when the data is in a single DICOM file.
I reached out to our school’s imaging center, and they mentioned that for long scans, they can only provide data in “dicom_classic” format.
I’m wondering:
- Why might
dcm2bidsintroduce artifacts thatdcm2niixdoes not, even though dcm2bids uses dcm2niix under the hood? I noticed that dcm2bids runs dcm2niix with the following info:Running dcm2niix -b y -ba y -z y -f '%3s_%f_%p_%t' -o .... Could it be that some of these parameters are causing the issue? - What is the best and easiest solution or workaround for us? One idea I had was to use
dcm2bidsto organize the data into BIDS format and then usedcm2niixseparately to convert the functional data to the correct NIfTI files and replace them. But I am wondering if we have a better option. Moreover, I also noticed that dcm2bids has a--skip_dcm2niix`` option, which seems like it could skip the DICOM conversion, but it seems this option is not available in the older version of dcm2bids (we are currently using version 2.1.6).
Any insights or suggestions would be greatly appreciated!
Yanbin