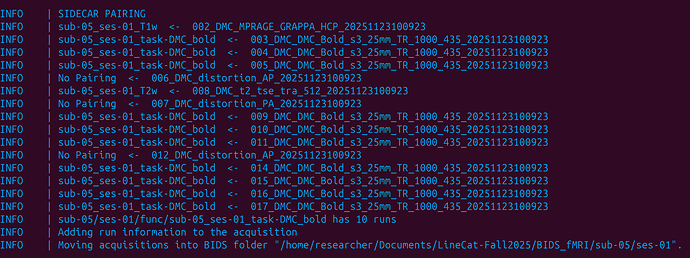
Summary of what happened:
I have two epi field maps, one AP and one PA, that I’d like to use for distortion correction on functional images. When running dcm2bids, I get a “No Pairing” error for these sequences. The json file I’m using is pasted below (using only one functional run for brevity). I’ve tried a couple of different approaches from different threads on this site but keep running into the same issue.
Command used (and if a helper script was used, a link to the helper script or the command generated):
"id": "DMC-run-10",
"datatype": "func",
"suffix": "bold",
"custom_entities": "task-DMC",
"criteria": {
"SidecarFilename": "017*"
},
"sidecar_changes": {
"TaskName": "DMC"
}
},
{
"datatype": "fmap",
"suffix": "epi",
"criteria": {
"SeriesDescription": "distortion_AP",
"dir" : "AP",
"PhaseEncodingDirection": "j-",
"SidecarFilename": "006*"
},
"sidecar_changes": {
"TaskName": "DMC",
"IntendedFor": [
"DMC-run-01",
"DMC-run-02",
"DMC-run-03",
"DMC-run-04",
"DMC-run-05",
"DMC-run-06",
"DMC-run-07",
"DMC-run-08",
"DMC-run-09",
"DMC-run-10"
]
}
},
{
"datatype": "fmap",
"suffix": "epi",
"criteria": {
"SeriesDescription": "distortion_PA",
"dir" : "PA",
"PhaseEncodingDirection": "j",
"SidecarFilename": "007*"
},
"sidecar_changes": {
"TaskName": "DMC",
"IntendedFor": [
"DMC-run-01",
"DMC-run-02",
"DMC-run-03",
"DMC-run-04",
"DMC-run-05",
"DMC-run-06",
"DMC-run-07",
"DMC-run-08",
"DMC-run-09",
"DMC-run-10"
]
}
}
Version:
3.2.0
Environment (Docker, Singularity / Apptainer, custom installation):
Data formatted according to a validatable standard? Please provide the output of the validator:
PASTE VALIDATOR OUTPUT HERE
Relevant log outputs (up to 20 lines):
PASTE LOG OUTPUT HERE
Screenshots / relevant information:
Here’s a screenshot of the dcm2bids output: