Hello everyone!
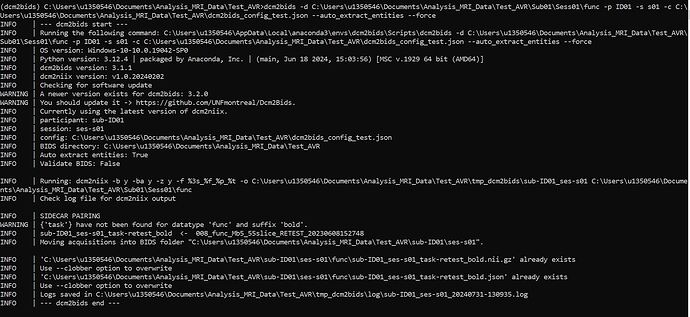
I’m trying to figure out how to convert my fMRI DCM files to BIDS and I’m getting the following warning: {‘task’} have not been found for datatype ‘func’ and suffix ‘bold’. I know that others have posted about this, but I do have the custom_entities in here and it’s still not working. Below you can find the content of my configuration file. I would appreciate any tips on how to fix this! ![]()
{
"descriptions": [
{
"datatype": "func",
"suffix": "bold",
"custom_entities": "task-retest",
"criteria": {
"ProtocolName": "Mb5_55slice_RETEST"
},
"sidecarChanges": {
"TaskName": "task-retest"
}
}
]
}