I am trying to denoise BOLD by regressing out the effects of CSF,WM and motion. I have preprocessed my data entirely in SPM and right now I have the smoothed images as well as a design matrix (SPM.mat) with regressors of interest as well as the motion and other nuisance regressors. If I am extracting BOLD from the SPM.mat does it mean my data is already denoised? If not what best way can I denoise my BOLD with these preprocessed files and not repeat the preprocessing entirely in fmriprep or etc? Thank you.
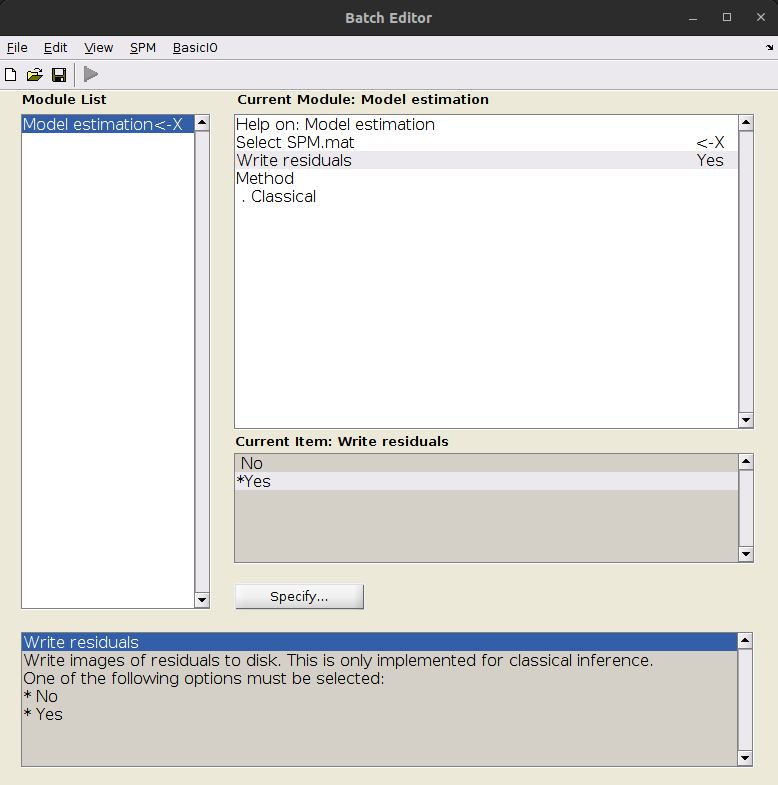
if you want to generate “denoised” time series you need to create a GLM where the design matric only includes your motion and other regressors of no interest and and ask SPM to ouput the residuals during model estimation
This should dump one 3D nifti image for each time point in your entire model, in the same folder as your SPM.mat.
You should be able to reconstuct your denoised time series from this.
Thank you so so much for your quick reply… I tried out so many other things like CONN, GLMsingle, and a few other scripts I found out there but none was working… I was stuck on this step for quiet a while, and since I wanted to compute BOLD variability this was a crucial step that I couldn’t skip… Thank you so so much… It worked!!