Dear TDT experts,
How can the design matrix be specified for two designs with two different sets within the same analysis?
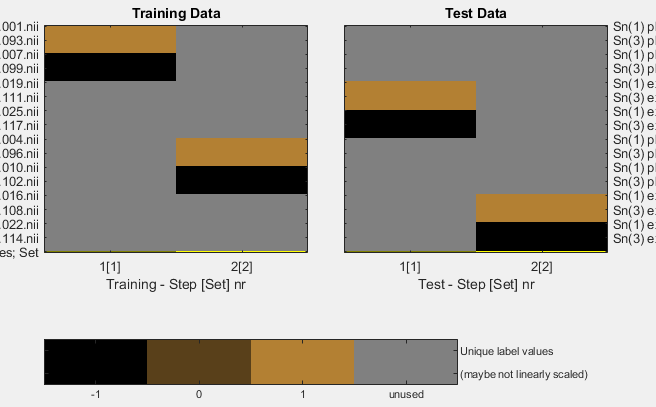
I’m talking about what Figure 3F from the TDT paper shows (figure caption: “Example design for two leave-one-run-out designs with two different sets, in the same decoding analysis. The results are reported combined or separately for each set which can speed-up decoding.”). The figure describes a cross-validation design for two sets, is it possible to perform a cross-classification with two sets?
The idea is to have (in the same analysis):
- design 1: training on labelname1classA labelname1classB, testing on labelname2classA labelname2classB,
- design 2: training on labelname1classC labelname1classD, testing on labelname2classC labelname2classD.
When trying:
cfg = decoding_describe_data(cfg,...
{labelname1classA labelname1classB labelname1classC labelname1classD ...
labelname2classA labelname2classB labelname2classC labelname2classD},...
[1 2 3 4 -1 -2 -3 -4],...
regressor_names,...
beta_loc,...
[1 1 2 2 -1 -1 -2 -2]);
I got the error:
Wrong number of labels in cfg.files.xclass. Cross classification needs exactly one training and one test set.
Hence I was wondering if 2-set cross-classification is allowed by the toolbox, or maybe the design is specified differently?
In addition, where/how can we define the results to be reported combined or separately? And related to that, where can we find more information on how the results from a two-set analysis are combined?
I appreciate any lights you can provide. Thanks in advance!
Ricardo.