Hi all,
If we run FMRIPREP twice on the same data, we get different results. That’s well known, and such variability can be avoided with the use of options such as --random-seed 0, --skull-strip-fixed-seed, --nthreads 1, and --omp-nthreads 1.
However, there is no obvious way to choose one seed over another in --random-seed, and for the analyses we are running, the choice of the seed causes relevant, non-ignorable differences in the final results.
Before we establish a procedure that will run FMRIPREP many times to cancel out such variability, we’d like to understand what causes it. We thought ANTs could be the culprit but we also observe temporal variability in the *_desc-preproc_bold.nii.gz, which rules ANTs out since registration would affect all volumes equally. We also thought of ICA-AROMA but the latter should (so we understand) not affect the *_desc-preproc_bold.nii.gz files, just produce covariates in the *_desc-confounds_timeseries.tsv file.
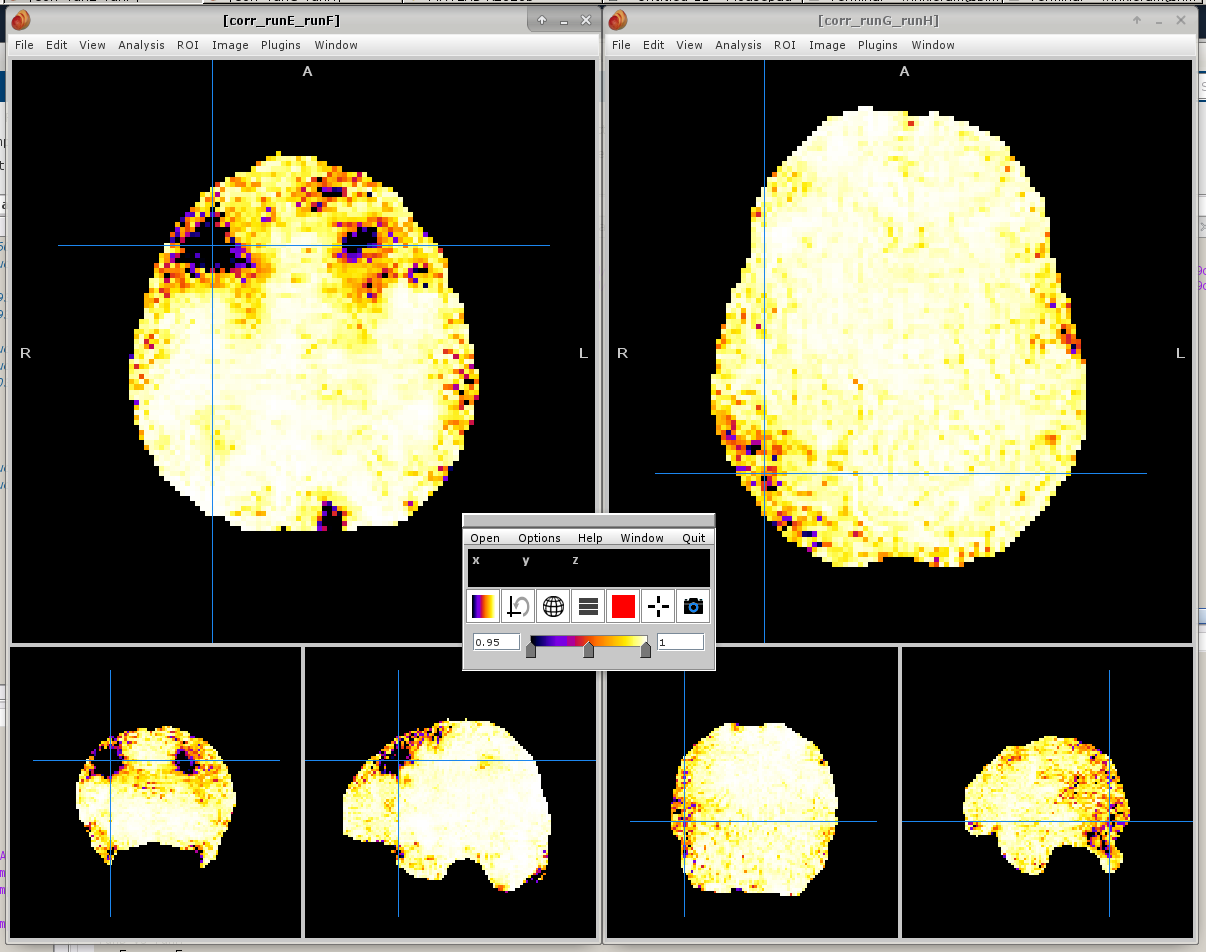
The screenshot below shows the maps of the correlations of the time series of pairs of runs of FMRIPREP. That is, for every voxel from *_desc-preproc_bold.nii.gz file, we correlate the time series of that voxel with the homologous voxel in the *_desc-preproc_bold.nii.gz from a different run of FMRIPREP (that presumably used a different random seed). Note that the variability isn’t random, having some spatial pattern. Note also that it isn’t negligible. While we’ve set the colorbar between 0.95 and 1, we have correlations here as low as 0.27 in some places – and have seen them even negative!
What could be causing this?
Thanks!
All the best,
Anderson
Just fyi, the calls to FMRIPREP were (the run names are in the headers of the windows in the screenshot):
# run E
fmriprep [REDACTED]/BIDS/AP2 ${TMPDIR}/s9900.out participant --participant_label s9900 -w ${TMPDIR}/s9900.wrk --notrack --output-space MNI152NLin2009cAsym:res-2 fsaverage --nthreads 1 --omp-nthreads 1 --skip_bids_validation --use-aroma
# run F
fmriprep [REDACTED]/BIDS/AP2 ${TMPDIR}/s9900.out participant --participant_label s9900 -w ${TMPDIR}/s9900.wrk --notrack --output-space MNI152NLin2009cAsym:res-2 fsaverage --nthreads 1 --omp-nthreads 1 --skip_bids_validation --use-aroma
# run G
fmriprep [REDACTED]/BIDS/AP2 ${TMPDIR}/s9900.out participant --participant_label s9900 -w ${TMPDIR}/s9900.wrk --notrack --output-space MNI152NLin2009cAsym:res-2 fsaverage --nthreads 1 --omp-nthreads 1 --skip_bids_validation
# run H
fmriprep [REDACTED]/BIDS/AP2 ${TMPDIR}/s9900.out participant --participant_label s9900 -w ${TMPDIR}/s9900.wrk --notrack --output-space MNI152NLin2009cAsym:res-2 fsaverage --nthreads 1 --omp-nthreads 1 --skip_bids_validation