Hi,
Summary of what happened:
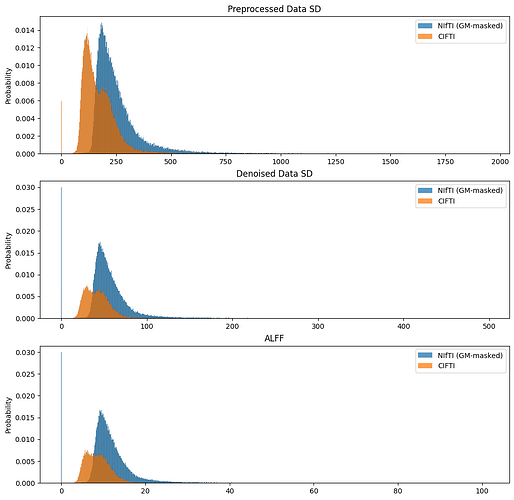
Large differences between ALFF values in the hippocampus obtained from a CIFTI output (hippocampal ALFF values still in volumetric space) and the ones obtained from the NIFTI whole brain output.
Setup:
- Data: ABCD dataset, downloaded with Datalad.
- Container: Singularity container for XCP-D.
- Version: XCP-D v0.10.0rc3
Description of the problem
A colleague ran the XCP-D pipeline with the CIFTI output. As we decided to share the resources, I extracted the volumetric hippocampal ALFF values from
*space-fsLR_den-91k_stat-alff_boldmap.dscalar.nii
with the command
wb_command -cifti-separate sub-XXX_ses-baselineYear1Arm1_task-rest_run-02_space-fsLR_den-91k_stat-alff_boldmap.dscalar.nii COLUMN -volume HIPPOCAMPUS_LEFT hippocampus_L.nii.gz
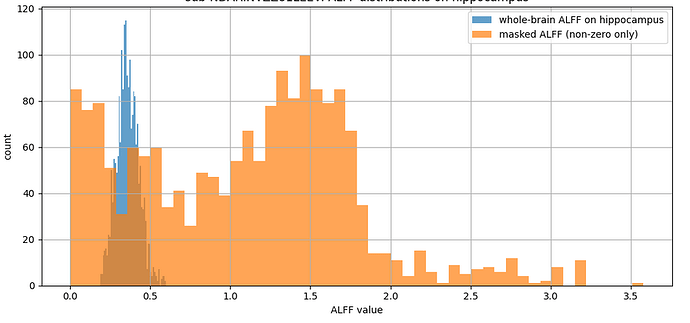
However, due to misalignment with the current hippocampal surface that I have (after adjusting for resolution and space), we decided to re-run the pipeline but this time to get the volumetric ALFF values for the whole brain. To our surprise, the ALFF values (in blue in Fig.1) of the hippocampus were much lower than the ones obtained with the hippocampal mask (in orange).
Fig. 1
We were wondering if the calculations may change depending on the output format, especially when considering allocortical and subcortical structures. The hippocampus mask extracted from the CIFTI file seems to be in the right place when plotted on the whole brain ALFF NIFTI file.
Excerpt of my colleague’s code:
# Apply transformation if output mask does not already exist
if [ ! -f "$output_mask" ]; then
echo "Applying transformation..."
antsApplyTransforms -d 3 -i "$mask_file" -r "$tpl_file" -n NearestNeighbor \
-t "$transform_file" -o "$output_mask" || {
echo "Error applying transformation for $mask_file"
missing_files_errors+=("$sub: Error applying transformation for $mask_file")
continue
}
echo "Transformation applied successfully!"
else
echo "Mask transformation output already exists for $mask_file"
fi
# Check if required BOLD data exists in fsLR space
fsLR_bold_files=$(find "$subject_dir/ses-baselineYear1Arm1/func/" -name "*_space-fsLR_den-91k_bold.dtseries.nii" 2>/dev/null)
if [ -z "$fsLR_bold_files" ]; then
echo "No BOLD data found in fsLR space for $sub. Skipping XCP-D processing."
missing_files_errors+=("$sub: No BOLD data in fsLR space")
else
# Run XCP-D post-processing pipeline
echo "Running XCP-D pipeline"
singularity run \
-B /data/ABCD:/data/ABCD \
--cleanenv /data/xcp_d-0.10.1.sif \
/data/ABCD/ABCD_fMRIprep/fmriprep \
/data/ABCD/XCP-D_output \
participant \
--mode 'none' \
--participant-label "${sub#sub-}" \
--bids-filter-file /data/ABCD/bids_filter_file.json \
--nprocs 36 \
--input-type 'fmriprep' \
--file-format 'cifti' \
--dummy-scans 'auto' \
--despike 'y' \
--nuisance-regressors /data/ABCD/custom_confounds_24P_csf_wm.yaml \
--fd-thresh 0.3 \
--output-type 'censored' \
--combine-runs 'n' \
--smoothing 6 \
--motion-filter-type 'none' \
--head-radius 50 \
--lower-bpf 0.01 \
--upper-bpf 0.08 \
--bpf-order 2 \
--min-time 240 \
--atlases '4S456Parcels' \
--min-coverage 0.5 \
--create-matrices 240 all \
--work-dir /data/ABCD/work \
--warp-surfaces-native2std 'n' \
--abcc-qc 'n' \
--linc-qc 'y' \
where
mask_file = {sub}_ses-baselineYear1Arm1"*desc-brain_mask.nii.gz;
transform_file = {sub}_ses-baselineYear1Arm1"*from-T1w_to-MNI152NLin6Asym_mode-image_xfm.h5
output_mask = {sub}_ses-baselineYear1Arm1_space-MNI152NLin6Asym_desc-brain_mask.nii.gz
tpl_file = /data/tpl-MNI152NLin6Asym/tpl-MNI152NLin6Asym_res-01_T1w.nii.gz
Excerpt of my code with alterations to return whole-brain volumetric ALFF values:
output_mask="${sub}_ses-baselineYear1Arm1_space-MNI152NLin6Asym_desc-brain_mask.nii.gz"
tpl_file="/data/pt_02983/data/ALFF/test/test2/tpl-MNI152NLin6Asym/tpl-MNI152NLin6Asym_res-02_T1w.nii.gz"
# pull ants container if needed
if [ ! -f /data/software/containers/ants_2.5.3.sif ]; then
echo "Pulling ANTs container..."
mkdir -p /data/software/containers
singularity pull /data/software/containers/ants_2.5.3.sif docker://antsx/ants:2.5.3
fi
echo "Running antsApplyTransforms for anatomical brain mask..."
singularity exec \
--cleanenv \
-B /data:/data \
/data/software/containers/ants_2.5.3.sif \
antsApplyTransforms -d 3 -v 1 \
-i "$mask_file" \
-r "$tpl_file" \
-n NearestNeighbor \
-t "$transform_file" \
-o "$output_mask" || {
echo "Error applying transformation for $mask_file"
continue
}
echo "Transformation applied successfully!"
# now handle the functional files
cd ../func/ || {
echo "Missing functional directory for $sub"
missing_files_errors+=("$sub: Missing functional directory")
continue
}
for bold_file in *task-rest_run-*_space-MNI152NLin6Asym_desc-smoothAROMAnonaggr_bold.nii.gz; do
bold_file=$(echo "$bold_file" | sed 's/"//g;s/@$//')
if [ -f "$bold_file" ]; then
symlink_file="${bold_file/_desc-smoothAROMAnonaggr/_desc-preproc}"
if [ ! -f "$symlink_file" ]; then
ln -s "$bold_file" "$symlink_file"
fi
boldref_file="${bold_file/_desc-smoothAROMAnonaggr_bold/_boldref}"
if [ ! -f "$boldref_file" ]; then
fslroi "$bold_file" "$boldref_file" 0 1
fi
# extract run id
run=$(echo "$bold_file" | grep -o "run-[0-9][0-9]")
# define run-specific anatomical mask symlink name
anatomical_mask_run="${sub}_ses-baselineYear1Arm1_task-rest_${run}_space-MNI152NLin6Asym_desc-brain_mask.nii.gz"
if [ ! -f "$anatomical_mask_run" ]; then
ln -s "$(realpath ../anat/${output_mask})" "$anatomical_mask_run"
echo "Created run-specific mask: $anatomical_mask_run"
fi
fi
done
# Run XCP-D post-processing pipeline
echo "Running XCP-D pipeline"
singularity run \
-B /data/:/data \
--cleanenv /data/xcp_d-0.10.0rc3.simg \
/data/ABCD/ABCD_fMRIprep/fmriprep \
/data/ALFF/test/test2/XCP-D_output/ \
participant \
--mode 'none' \
--participant-label "${sub#sub-}" \
--bids-filter-file /data/ALFF/test/test2/bids_filter_file.json \
--nprocs 36 \
--input-type 'fmriprep' \
--file-format 'nifti' \
--dummy-scans 'auto' \
--despike 'y' \
--nuisance-regressors /data/ALFF/test/test2/custom_confounds_24P_csf_wm.yaml \
--fd-thresh 0.3 \
--output-type 'censored' \
--combine-runs 'n' \
--smoothing 6 \
--motion-filter-type 'none' \
--head-radius 50 \
--lower-bpf 0.01 \
--upper-bpf 0.08 \
--bpf-order 2 \
--min-time 240 \
--skip-parcellation \
--create-matrices 240 all \
--work-dir /data/ALFF/test/test2/work \
--warp-surfaces-native2std 'n' \
--abcc-qc 'n' \
--linc-qc 'y' \
Thank you very much in advance! (:
Best,
Mylla