I’m experimenting with recognizing fibers on tractograms by using machine learning. My goal is to automate bundle extraction from a patient. It looks very promising so far - it doesn’t require any tweaking unlike RecoBundles from DIPY.
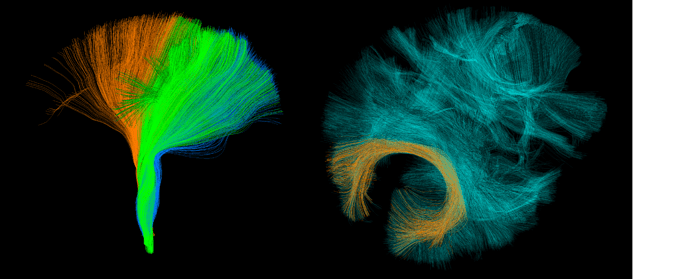
On the first pic you see FPT, CST and PPT bundles. They mix up, but still somewhat separable because their lower tips go to different areas. On the second pic you see Forceps Major and CC as it is labeled in HCP842 atlas supplied with DIPY.
-
Why there’s plenty of fibers in CC very close and similar to Forceps Major fibers? Is it just from poor labeling or there’s something I don’t understand?
-
From the whole tractography RecoBundles selects fibers that are close enough (by threshold) to model fibers, and doesn’t consider models of other bundles. Thus some fibers could be RecoBundled as multiple bundles at the same time - e.g. FPT and CST. Why does RecoBundles use just distances and thresholds and doesn’t compare fibers against multiple bundles at once? Is there any reason why fibers should be compared in this exact way?
Hi @mmatrosov
1- The original HCP842 atlas has only one Copus Collusum bundle provided as CC.trk. For our paper Bundle Analytics, we divided CC.trk into three parts, Forceps Major, Forceps Minor, and Copus Collusum Middle part. The HCP842 atlas provided by DIPY is a curated version of the original one. There are also some differences in the Arcuate Fasciculus and Uncinate Fasiculus bundles in two atlases. We kept the original entire CC bundle along with its three subset bundles just in case someone prefers to extract the entire CC. This is the reason you see an overlap in the CC and Forceps Major fibers.
2- Recobundles is designed that way to extract streamlines that look like the model bundle streamlines from the input tractogram. Here we provide this curated/altered HCP842 atlas in DIPY but say someone wants to use their own atlas and they have just one model bundle, they don’t need to have all the bundles in order to compare if streamline should be labeled as A or B, C, etc. You do make a good point though that sometimes we can end up labeling the same streamline as part of two bundles. That really depends on the model bundle you provide to RecoBundles. Also, there isn’t one golden standard definition of bundles which could be one reason why we see discrepancies in some model bundles. See this paper on variability in tract extraction methods https://www.biorxiv.org/content/10.1101/2020.10.07.321083v3
Hope this helps!
Bramsh
Thank you so much @bramsh!
-
I mean there are 6 times more CC fibers at the same place where FMaj, FMin and Mid, some of which are exactly the same, but most of them aren’t.
All 365 FMaj fibers (orange), 2236 fibers from CC similar to FMaj (blue):
So the questions is: are these additional 2236-365=1871 fibers also should be labeled as FMaj, or they are substantially different in a way that I don’t understand (e.g. not highlighted in fMRI tests or smth)?
-
I see, thanks again! As far as I understand this also indirectly answers my first question. Anyway, would you please confirm my assumption?
Hi @mmatrosov
Our neuroanatomist dissected the CC bundle into three parts, Forceps Major, Minor, and Mid. If you prefer a slightly different definition of any of these bundles, you can provide your own model bundle to RecoBundles and it would work. Again it is not necessary to use the HCP842 atlas. You can use your own atlas or any other atlas. Playing with some thresholds might also help in getting desired thinner or denser bundles DIPY : Docs 1.4.1. - Automatic Fiber Bundle Extraction with RecoBundles
1 Like