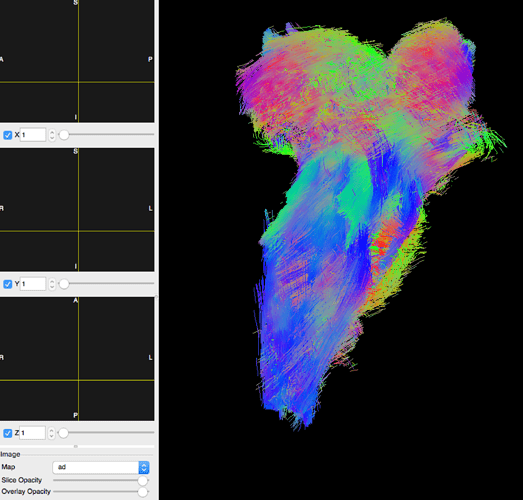
I’m using DIPY’s workflows to estimate DTI and CSD and create streamlines from these inputs. Each node is creating the appropriate inputs, but the streamlines don’t appear in the diffusion image space, despite taking DTI and CSD outputs as inputs: (full code below)
wf.connect(recon_dmri, 'out_pam', tracker, 'pam_files')
wf.connect(recon_dti, 'out_fa', tracker, 'stopping_files')
wf.connect(masker, 'mask_filename', tracker, 'seeding_files')
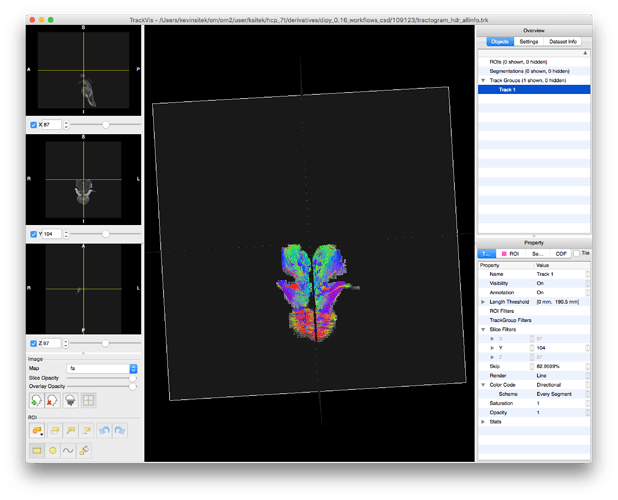
After loading the streamlines in TrackVis and importing the MD image created in the DTI workflow node, the MD image fails to appear: (this also happens loading both tractogram.trk and md.nii.gz in dipy_horizon)
Viewing the MD and other DTI derivatives on their own is fine, leading me to question the space mismatch during the tractogram generation process.
And actually when I first load the tractogram in TrackVis, it’s hyper-zoomed in, doubling down my space thoughts.
Any ideas how to fix the tractogram space issue? @Ariel_Rokem
Here’s the full code: