Discrepancies in t2smap Values between Python-wrapper and cmd-line TEDANA
Hello Tedana users!
I am new to Tedana and would appreciate your help!
I attempted to reproduce the T2starmap.nii.gz from fmriprep (v23.2.3) t2smap_node work folder with the Pypi Tedana package (for comparability, I used Tedana v23.0.2). I ran both the cmd-line and python wrapper. For inputs, I used the same time series and mask files as the fmriprep’s t2starmap command (found in t2smap_node folder) used.
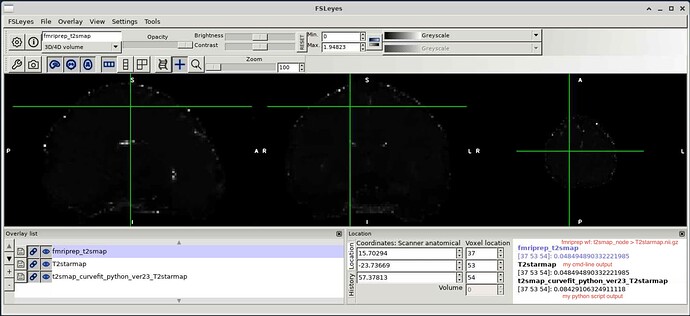
However, only the cmd-line script was able to reproduce the same t2smap. The python script resulted in some noticeable t2s value discrepancies across the brain.
I wonder what caused the discrepancy and which output should I use.
Thank you!
Command used:
cmd-line script, copied from fmriprep’s t2smap_node work folder:
conda activate fmri_env # where I pip installed tedana==23.0.2 at
t2smap -d ${studyFolder}/fmriprep/sub-000/fmriprepWork/fmriprep_23_2_wf/sub_000_wf/bold_ses_2_task_ME3_run_1_echo_1_wf/bold_native_wf/_echoidx_0/boldref_bold/sub-000_ses-2_task-ME3_run-1_echo-1_bold_tshift_xformresampled.nii.gz \
${studyFolder}/fmriprep/sub-000/fmriprepWork/fmriprep_23_2_wf/sub_000_wf/bold_ses_2_task_ME3_run_1_echo_1_wf/bold_native_wf/_echoidx_1/boldref_bold/sub-000_ses-2_task-ME3_run-1_echo-2_bold_tshift_xformresampled.nii.gz \
${studyFolder}/fmriprep/sub-000/fmriprepWork/fmriprep_23_2_wf/sub_000_wf/bold_ses_2_task_ME3_run_1_echo_1_wf/bold_native_wf/_echoidx_2/boldref_bold/sub-000_ses-2_task-ME3_run-1_echo-3_bold_tshift_xformresampled.nii.gz \
-e 15.4 37.220000000000006 59.02 \
--mask ${studyFolder}/fmriprep/sub-000/fmriprepWork/fmriprep_23_2_wf/sub_000_wf/bold_ses_2_task_ME3_run_1_echo_1_wf/bold_native_wf/bold_t2smap_wf/dilate_mask/dilated_mask.nii.gz \
--fittype curvefit
python script:
# jupyter notebook operated in the same conda environment as the cmd-line script
TE1_bold = studyFolder+'/fmriprep/sub-000/fmriprepWork/fmriprep_23_2_wf/sub_000_wf/bold_ses_2_task_ME3_run_1_echo_1_wf/bold_native_wf/_echoidx_0/boldref_bold/sub-000_ses-2_task-ME3_run-1_echo-1_bold_tshift_xformresampled.nii.gz'
TE2_bold = studyFolder+'/fmriprep/sub-000/fmriprepWork/fmriprep_23_2_wf/sub_000_wf/bold_ses_2_task_ME3_run_1_echo_1_wf/bold_native_wf/_echoidx_1/boldref_bold/sub-000_ses-2_task-ME3_run-1_echo-2_bold_tshift_xformresampled.nii.gz'
TE3_bold = studyFolder+'/fmriprep/sub-000/fmriprepWork/fmriprep_23_2_wf/sub_000_wf/bold_ses_2_task_ME3_run_1_echo_1_wf/bold_native_wf/_echoidx_2/boldref_bold/sub-000_ses-2_task-ME3_run-1_echo-3_bold_tshift_xformresampled.nii.gz'
TE1=15.4
TE2=37.220000000000006
TE3=59.02
dil_mask = studyFolder+'/fmriprep/sub-000/fmriprepWork/fmriprep_23_2_wf/sub_000_wf/bold_ses_2_task_ME3_run_1_echo_1_wf/bold_native_wf/bold_t2smap_wf/dilate_mask/dilated_mask.nii.gz'
tedana.workflows.t2smap_workflow([TE1_bold, TE2_bold, TE2_bold],
[TE1, TE2, TE3],
out_dir=".",
mask=dil_mask,
prefix='t2smap_curvefit_python_ver23',
convention='bids',
fittype='curvefit',
fitmode='all',
combmode='t2s',
debug=False,
quiet=False)
The cmd-line output and fmriprep’s wf T2starmap.nii.gz are identical throughout the brain.
But the py-wrapper output is systematically different. I’m wondering if people have noticed this before and if they have any thoughts on why this is the case?
Version:
tedana 23.0.2
fmriprep 23.2.3
Environment (Docker, Singularity / Apptainer, custom installation):
my fmri_env conda environment:
Python 3.10.12
tedana 23.0.2
Screenshots / relevant information:
View cmd-line produced, py-wrapper produced and fmriprep’s T2starmap.nii.gz: