Hello,
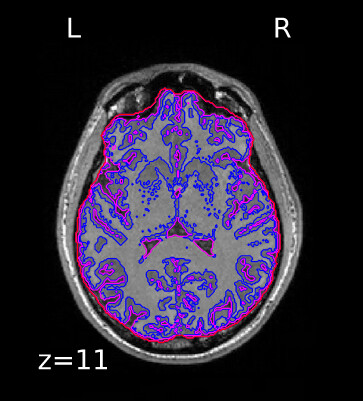
I want to create a report similar to what fMRIprep does. To start, I’d like to display tissue segmentation countours on T1 volume, like this example by fMRIprep. Do I use nilearn to do that?
Hello,
I want to create a report similar to what fMRIprep does. To start, I’d like to display tissue segmentation countours on T1 volume, like this example by fMRIprep. Do I use nilearn to do that?

Yes, you can, taking inspiration from 7. Plotting brain images - Nilearn
Note also the existence of GitHub - neurospin/pypreprocess: Preprocessing scripts for neuro imaging
HTH,
Bertrand
Thanks,
With Nilearn It doesn’t seem to be possible to display reconstructed white and pial surfaces. Is there any other visualization tool that can be used for that?
Freeview is the classical tool for that.
I’m not sure, but possibly Niivue can do it now.
Best,
Bertrand
Thanks! I know about Freeview, I was looking to have some outomatic report generation.
There are at least two ways to show these contour images of the segmentation. The first kind of display is a standard part of visualization in AFNI with the afni and suma GUIs “talking” to each other. The suma GUI shows surfaces, often from FreeSurfer, and the afni GUI shows the surface intersection on the volumetric slices. Here’s an example to get you started on how to automate that.
# start suma surface display
suma -spec std.141.FT_both.spec -sv FT_SurfVol.nii &
# start afni volumetric display
afni -niml -yesplugouts
# tell suma and afni to talk
DriveSuma -com viewer_cont -key 't'
# tell afni to pick specific datasets for underlay and overlay
plugout_drive -com 'SWITCH_SESSION A.afni' \
-com 'OPEN_WINDOW A.axialimage geom=600x600+416+44 \
ifrac=0.8 opacity=9' \
-com 'OPEN_WINDOW A.sagittalimage geom=+45+430 \
ifrac=0.8 opacity=9' \
-com 'SWITCH_UNDERLAY FT_SurfVol.nii' \
-com 'SWITCH_OVERLAY strip' \
-com 'SEE_OVERLAY +' \
-com 'SET_DICOM_XYZ -32 24 23' \
-com 'SAVE_JPEG axialimage Saved.jpg' \
-quit
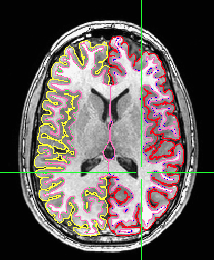
Another way is to compose your own volumetric dataset of contours - edges of the segmentation. You can do that by using the depth distances of each segmentation type and limiting that to just a rim.
3dDepthMap -input aseg.nii.gz -rimify 1 -prefix aseg_rim1.nii.gz
Save that with a similar afni-plugout_drive command as in the previous example or use an @chauffeur_afni command to let that do the driving.
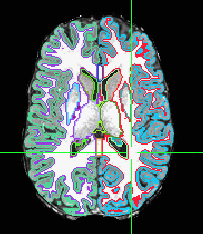
@chauffeur_afni
-ulay brain.nii.gz
-olay aseg_rim1.nii.gz
-pbar_posonly
-cbar “ROI_i256”
-func_range 256
-opacity 4
-prefix rimmed_brain
-montx 5 -monty 3
-set_xhairs OFF
-label_mode 1 -label_size 4
-do_clean
Thanks, I will try these out!
As an aside, I am a fan of the difference of gaussian (DoG) method for creating contours. Here are Python, Matlab and native executables for computing DoG. Nice features of DoG are that it enforces closed contours (like contour lines on an altitude map) and it works on 3D not 2D images, so the contour lines are coherent regardless of whether you view them on axial, sagittal or coronal slices.
MANAGED BY INCF