Summary of what happened:
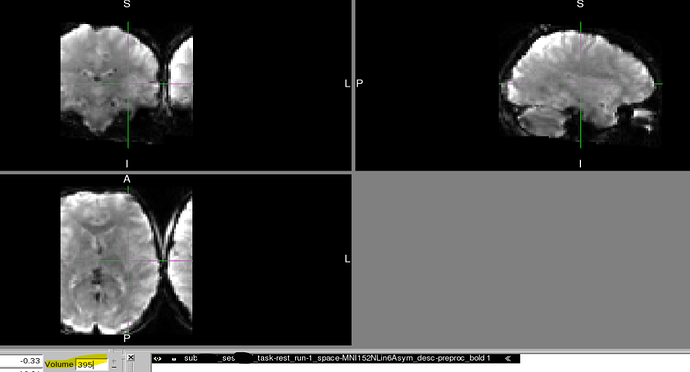
After transforming our T2* functional data into MNI152NLin6Asym space using fmriprep 23.2.1, our images became distorted, but this distortion only manifests midway through the time course. Such distortion did not exist at preceding preprocessing steps, such as normalization to the subject level T1w or within the raw data.
Command used (and if a helper script was used, a link to the helper script or the command generated):
singularity run --cleanenv -B /${TMPROOT}:/${TMPROOT} ${FMRIPREP} \
${BIDS_DIR} ${OUTPUT_DIR} participant \
--fs-license-file=${FS_LICENSE} \
--participant-label=${SUB} \
-w ${WORK_DIR} \
--cifti-output \
--output-spaces anat fsaverage MNI152NLin6Asym \
--ignore slicetiming \
--return-all-components
Version:
23.2.1
Environment (Docker, Singularity / Apptainer, custom installation):
Singularity
Data formatted according to a validatable standard? Please provide the output of the validator:
"We found 4 Warnings in your dataset."
(Warnings are known to team and are expected. Ex, missing sessions due to withdrawals.)
Relevant log outputs (up to 20 lines):
N/A
Screenshots / relevant information:
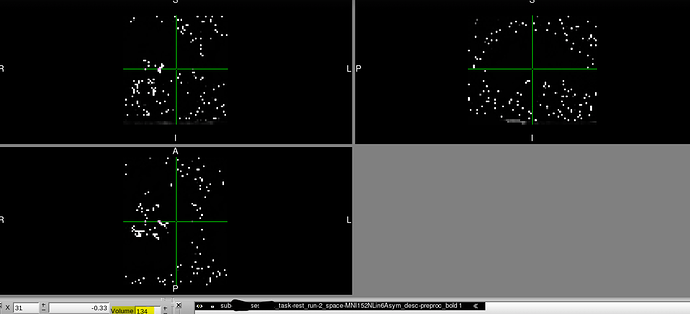
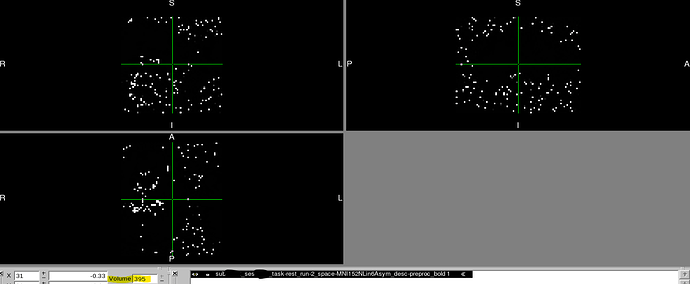
We recently discovered that at least two of our T2* functional scans became distorted only when transforming to MNI space (MNI152NLin6Asym) and only partway through their time course.
Basically:
- Raw data shows no similar distortion throughout its full time course.
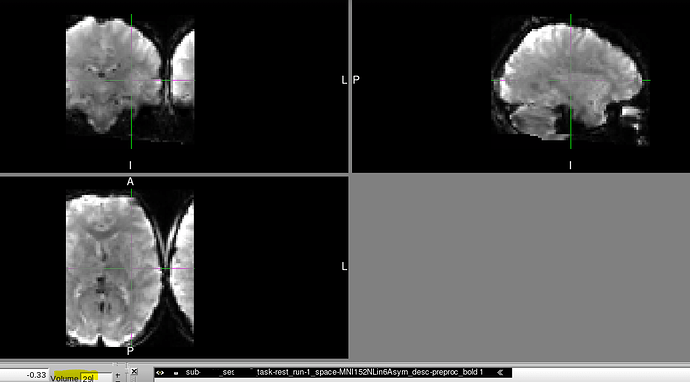
- Data normalized to the subject level T1w shows no similar distortion throughout its full time course.
- fMRIprep HTML QC report shows no similar distortion in any of its visualizations.
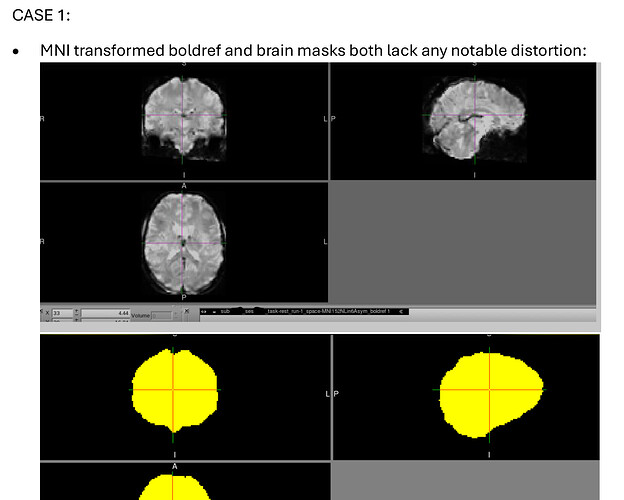
- fMRIprep’s MNI transformed boldref shows no similar distortion.
- fMRIprep’s MNI transformed brain mask shows no similar distortion.
- Beginning of MNI transformed time course shows no similar distortion.
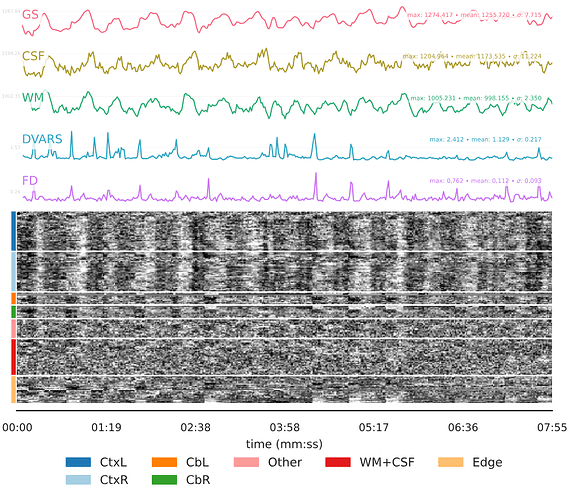
However, partway through the time course after transforming to MNI space, the image is distorted in dramatic and perplexing ways. It should also be noted that:
- Once the image becomes distorted, it remains this way for the rest of the sequence.
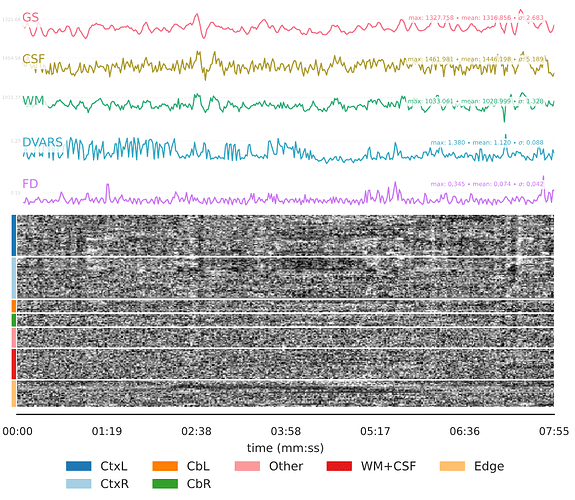
- There was no movement or other unusual event around the time when the sequence became distorted, as evidenced by all preceding steps appearing unaffected. Overall movement was also low in both cases with a mean FD of 0.112 for CASE 1 and 0.074 for CASE 2.
- Both individuals had a second run of the exact same sequence collected on the same day, and there were no such issues in that alternate run. Both individuals also collected another time point using the same sequences and there were no such issues, so it seems unlikely that these are subject-level phenomena.
These two examples were discovered inadvertently when running FEAT, so we are concerned that other scans could have been impacted by similar errors without us knowing, as we encountered no issues when running fmriprep itself. If anyone has any insight as to why this could be happening, we’d be very grateful.
Below, you can find screenshots of the two cases. Apologies also for the long image. New users are not permitted to embed multiple screenshots.