Hi experts and users
I have used this code to preprocess my fmri data:
fmriprep-docker /home/xxx/bids/ /home/xxx/bids/derivatives participant --participant-label fmri01 --fs-license-file /home/siiying/license.txt --omp-nthreads 25 --nprocs 25 -t ddt --use-aroma --fs-no-reconall
The *desc-preproc_bold.nii.gz data will be used in the following analysis. Do i need perform high-pass filtering, smooth, scale on the “*desc-preproc_bold.nii.gz”?
thanks
best regards
rujing
Hi @rujing and welcome to neurostars!
None of the steps that you described are done by fMRIprep, with the exception of smoothing which is only applied to AROMA outputs. So they are something you would have to apply yourself. Of note, including the cosine regressors from the confounds in your first level GLM will high-pass your data at 1/128 hz.
Also as a note, it is not recommmended to use the fs-no-reconall flag, as FreeSurfer reconstructions improve anatomical alignments to template spaces, among other benefits.
Best,
Steven
Hi @Steven
Thank you. I see. A further question is I have no B0 fieldmap. However i would like to perform distortion correction using some options, such as --use-syn-sdc and --force-syn. Is this recommended? I am new to fmriprep. AFNI tools were used in my previous work and no distortion correction was performed. So i would like to know whether these two options were used correctly.
Thanks
Happy New Year!
rujing
You can try use-syn-sdc and see if it works well for you and then decide it you want to use it. (force-syn is not needed usually to get it to work unless you have fieldmaps that would otherwise take precedence). SDC improves bold-to-T1 spatial correspondences, which is more important in some analyses than others.
Hi @Steven
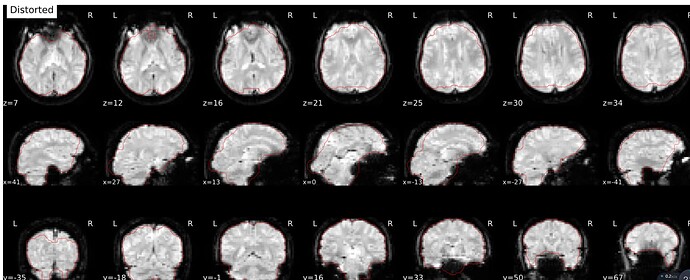
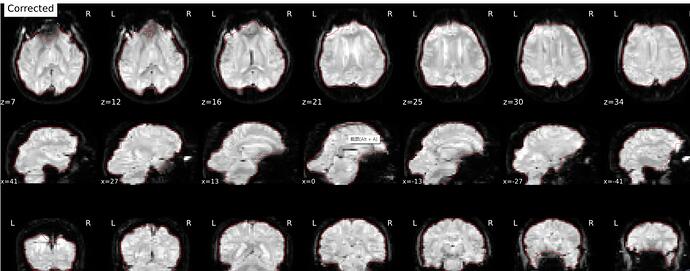
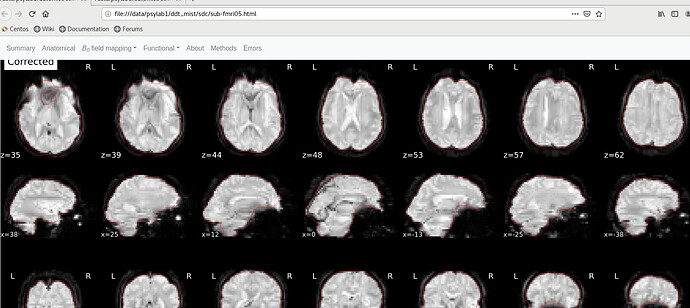
I have added use-syn-sdc in the code, and “About”, “Methods”, “Errors” in the sub*.html were uploaded in this message. I see No errors to report in “Errors”, and does this represent the use-syn-sdc option works well for me?
Thanks.
Best regards.
rujing
You should check the SDC report in the html file to see if the corrected EPI borders more closely line up with the T1 boundary (red outline)
Hi @Steven
In the SDC report in the html file, corrected version is well fit with the red outline, but the distorted version is not. So, does this mean that the use-syn-sdc works well in the data?
Thanks in advance.
Best regards.
rujing
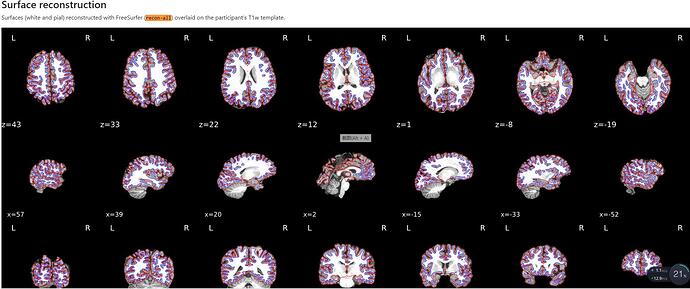
It looks like SDC is doing its job, but the T1 outline looks weird. Did you use FreeSurfer recon-all outputs?
Hi @Steven
This is my code: docker run --rm -v /home/psylab1/psylab1/ddt_mist/bids:/input -v /home/psylab1/psylab1/ddt_mist/bids/derivatives:/output -v /home/psylab1/psylab1/ddt_mist/license.txt:/license nipreps/fmriprep:22.1.0 /input /output participant --participant-label $sbname --fs-license-file /license --omp-nthreads 1 --nprocs 1 -t ddt --use-syn-sdc error --use-aroma
I think the code used the recon-all. The surface was uploaded in this message.
How does it look on other subjects? Maybe just this one subject has the weird distortion in the back of the brain.
Hi @rujing, I think that the read outline on those images is the outline of the brain mask calculated from the “corrected” EPI images, with not link with the anatomical images, which seems fine here. So it seems that SDC was applied, but wrongly, because the corrected image is less like a normal brain than the distorted image.
As @Steven mentioned, is it the case for all your subjects? I don’t think that your original bold image looks particularly heavily distorted, it is more an issue with how the fieldmap was estimated and applied during SDC using the experimental fieldmap-less method.
Which fmriprep version did you use? Different fmriprep versions leverage different SDCFlows versions for SDC and can give different results for the fieldmap-less SDC method (for other methods too!). If the fieldmap-less method does not provide better correction than this for your subjects (i.e. SDC outputs more distorded image that the original uncorrected one), then you would be better off not using SDC but using only the bold-to-t1 registration with 12 dot (--bold2t1w-dof 12 in fmriprep command line) to best align your bold image to the anatomical image when no SDC is done.
Of note, outside of FMRIPREP other fieldmap-less methods exist which can give other results. You can see these tools for instance:
Hi @Steven
Thanks very much for your help.
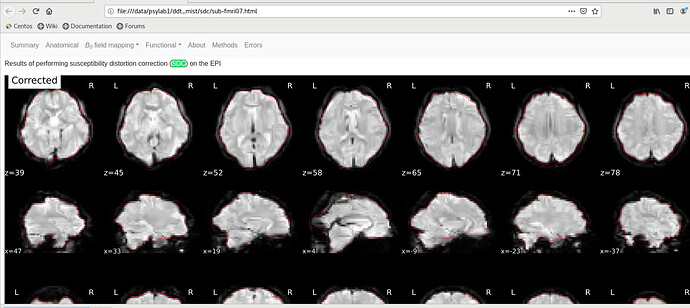
In the left subjects, I randomly selected two subjects. It seems that both subjects showed a weird red line boundary.
Hi @jsein
I used nipreps/fmriprep:22.1.0. This is my code: docker run --rm -v /home/psylab1/psylab1/ddt_mist/bids:/input -v /home/psylab1/psylab1/ddt_mist/bids/derivatives:/output -v /home/psylab1/psylab1/ddt_mist/license.txt:/license nipreps/fmriprep:22.1.0 /input /output participant --participant-label $sbname --fs-license-file /license --omp-nthreads 1 --nprocs 1 -t ddt --use-syn-sdc error --use-aroma
Thanks for your help.
rujing