I appreciate any advice you can offer here.
I have downloaded the gradient table from the HCP project from here (https://wiki.humanconnectome.org/display/PublicData/Gradient+Vector+Direction+table+for+HCP+3T+dMRI), and cleaned up the file to be a simple CSV with three columns representing the values from the gradient table (the b0 values are interleaved).
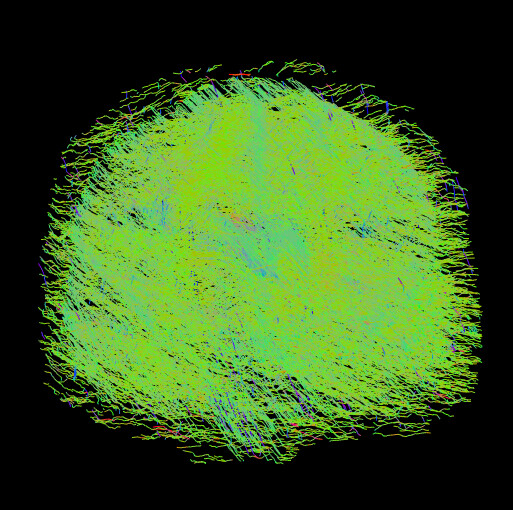
When I use Diffusion toolkit to perform reconstruction and tracking using a DTI imaging model, the tract file looks pretty good in trackvis with default settings. When I switch to HARDI/Q-ball, using the same gradient table, it is obviously wrong.
What am I doing wrong?