Downsampling is treacherous due to aliasing effects. Schumacher’s seminal chapter General Filtered Image Rescaling describes this issue as well as providing one elegant solution. I think there are three solutions to this problem:
- blur data to Nyquist limit prior to downsampling
- use aliasing-aware FFT.
- adapt kernel width to compensate for aliasing (Schumacher’s approach).
I suggest you look at the links in this previous question. Specifically, AFNI’s 3dresample does not employ an anti-aliasing filter. You can see this by running the Python code below to generate a 3D zone plate. and then downsampling the resulting image using 3dresample:
3dresample -input zoneplate.nii -prefix cu -dxyz 2.844444444 2.844444444 2.844444444 -rmode Cu
3dresample -input zoneplate.nii -prefix li -dxyz 2.844444444 2.844444444 2.844444444 -rmode Li
3dresample -input zoneplate.nii -prefix nn -dxyz 2.844444444 2.844444444 2.844444444 -rmode NN
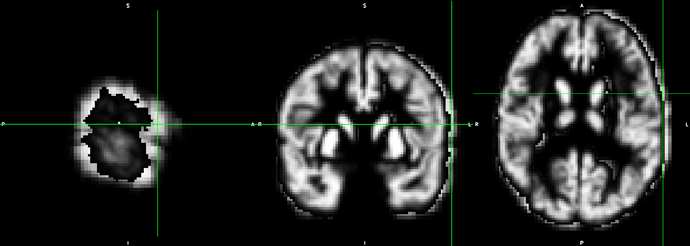
Below is the input image (left), 3dresample’s Cubic resampling (middle), and MRIcroGL’s Import/Tools/ResizeAndResample using anti-aliased Lanczos (right, using the method of Schumacher). The Matlab script nii_scale_dims would also anti-alias the data.

#!/usr/bin/env python3
# -*- coding: utf-8 -*-
# python3 d_speed_size.py : test speed/compression for folder 'corpus'
# python3 d_speed_size.py indir : test speed/compression for folder 'indir'
import math
import numpy as np
import nibabel as nib
nvox = 128
print("Creating 3D zone plate of with {} rows/columns/slices".format(nvox))
img = np.zeros((nvox, nvox, nvox))
center = (img.shape[0]/2.,img.shape[1]/2.,img.shape[2]/2.)
grid_x, grid_y,grid_z = np.mgrid[0:img.shape[0], 0:img.shape[1], 0:img.shape[2]]
grid_x = grid_x - center[0]
grid_y = grid_y - center[1]
grid_z = grid_z - center[2]
img = np.sqrt (np.square(grid_x) + np.square(grid_y) + np.square(grid_z))
img = np.reshape(img, (nvox, nvox, nvox))
header = nib.Nifti1Header()
affine = np.array([[1,0,0,-center[0]],[0,1,0,-center[1]],[0,0,1,-center[2]],[0,0,0,1] ])
nii = nib.Nifti1Image(img, affine, header)
nib.save(nii, 'distance.nii')
km = 0.7*math.pi
rm = max(center)
w = rm/10.0;
with np.nditer(img, op_flags=['readwrite']) as it:
for vox in it:
term1 = math.sin( (km * math.pow(vox,2)) / (2 * rm) )
term2 = 0.5*np.tanh((rm - vox)/w) + 0.5;
vox[...] = term1 * term2;
nii = nib.Nifti1Image(img, affine, header)
nib.save(nii, 'zoneplate.nii')