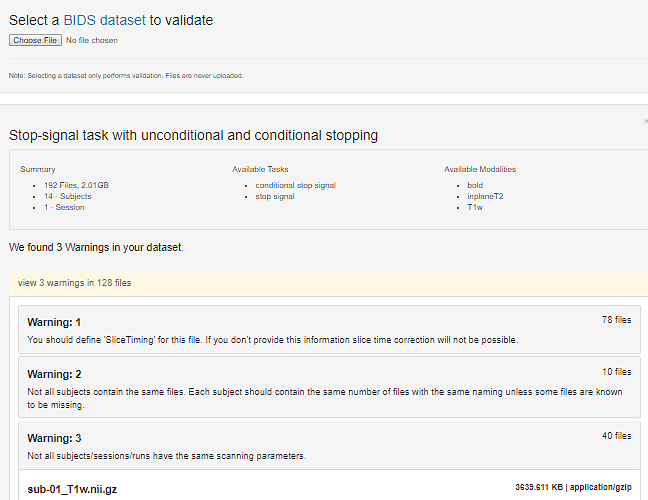
I am using the ds008 dataset from OpenfMRI.org as an example for some training. One of the tasks is to compare the NIfTI header information from the images to the information published.
From the published article cited at Openfmri.org,
The following appears in the section, Experiment 2: fMRI with the conditional stop-signal paradigm, subsection fMRI Data Aquisition,
The experiment was run using a 3T Siemens Allegra MRI scanner at the UCLA Ahmanson-Lovelace Brain Mapping Center. Each scanning run acquired 166 functional T2*-weighted echoplanar images (EPIs) [4 mm slice thickness; 33 slices; TR, 2 s; TE, 30 ms; flip angle, 90°; matrix, 64 × 64; field of view (FOV), 200; in-plane resolution, 3.125 mm]. The first two volumes in each run were discarded to allow for T1 equilibrium effects.
When I run
$ fslinfo sub-01_task-conditionalstopsignal_run-01_bold.nii.gz | grep
^dim
dim1 64
dim2 64
dim3 30
dim4 176
That looks to me like it should correspond to a 64 x 64 matrix, but with 30 slices (not the 33 reported) and that there are 176 slices in the volume, not the 164 (166-2) reported volumes acquired?
Similarly, they report for the T2 MP-RAGE images
The MP-RAGE acquisition parameters were as follows: TR, 2.3; TE, 2.1; FOV, 256; matrix, 192 × 192; sagittal plane; slice thickness, 1 mm; 160 slices.
I would anticipate from that the first and second dims would be 192 and the third 160, but the image header contains
$ fslinfo sub-01_inplaneT2.nii.gz | grep ^dim
dim1 128
dim2 128
dim3 30
dim4 1
When I run fslinfo on the T1 weighted image, I get
$ sub-01_T1w.nii.gz | grep ^dim
$ dim1 176
$ dim2 192
$ dim3 192
$ dim4 1
Am I missing something really obvious that would be apparent to an experienced analyst?
Thanks, – bennet