itreves
September 8, 2024, 8:35pm
1
Hi, I’m trying to run qsiprep and it isn’t registering my dwi images, “no dwi images found”. Do I need to convert this data? mrconvert?
PASTE CODE HERE
PUT VERSION HERE
PUT ENVIRONMENT HERE
PASTE VALIDATOR OUTPUT HERE
PASTE LOG OUTPUT HERE
Steven
September 8, 2024, 8:51pm
2
Hi @itreves and welcome to neurostars!
In the future please use the Software Support post category which prompts you to organize your post with important information we can use to help. You can see I reorganized your post accordingly.
Your data must be in BIDS format. See more here (Welcome to the BIDS Starter Kit — BIDS starter kit ).
Best,
1 Like
itreves
September 8, 2024, 9:08pm
3
Thanks Steven. Is this the correct bids format?
In addition, what should I do with the additional files with the opposite phase encoding ¶ and other series numbers (10,18,23), and what do you think the 79, 80 refer to?
Steven
September 8, 2024, 9:26pm
4
Hi @itreves ,
No that is not correct. Please refer to the specification: Magnetic Resonance Imaging - Brain Imaging Data Structure 1.10.0
They should also be in the DWI folder in BIDS. Unless they are just a single RPE b0, in which case that goes into the fmap folder.
The 79 and 80 refer to how many directions are acquired in the image (number of volumes).
For future posts, it is preferred to not use screenshots and instead use the tree command to return directory structures, formatted as code using the </> button in the text editor or surrounding the text by tick marks so it looks nice
like this.
Best,
itreves
September 9, 2024, 1:43pm
6
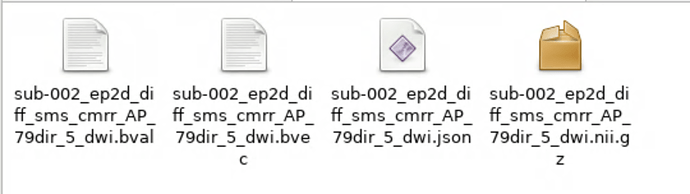
Here is my second attempt. There are no fieldmaps for the dwi. I’m indicating the number of volumes as acquisition, and direction for phase encoding.
.
├── sub-002_ses-1_acq-79_dir-AP_dwi.bval
├── sub-002_ses-1_acq-79_dir-AP_dwi.bvec
├── sub-002_ses-1_acq-79_dir-AP_dwi.json
├── sub-002_ses-1_acq-79_dir-AP_dwi.nii.gz
├── sub-002_ses-1_acq-79_dir-PA_dwi.bval
├── sub-002_ses-1_acq-79_dir-PA_dwi.bvec
├── sub-002_ses-1_acq-79_dir-PA_dwi.json
├── sub-002_ses-1_acq-79_dir-PA_dwi.nii.gz
├── sub-002_ses-1_acq-80_dir-AP_dwi.bval
├── sub-002_ses-1_acq-80_dir-AP_dwi.bvec
├── sub-002_ses-1_acq-80_dir-AP_dwi.json
├── sub-002_ses-1_acq-80_dir-AP_dwi.nii.gz
├── sub-002_ses-1_acq-80_dir-PA_dwi.bval
├── sub-002_ses-1_acq-80_dir-PA_dwi.bvec
├── sub-002_ses-1_acq-80_dir-PA_dwi.json
└── sub-002_ses-1_acq-80_dir-PA_dwi.nii.gz
Steven
September 9, 2024, 1:52pm
7
Hi @itreves ,
Looks better! But the BIDS validator will be the real test, as it considers the overall organization of data, file names, and metadata. It should be run internally by qsiprep if you do not --skip-bids-validation. Otherwise you can run it independent of qsiprep. (BIDS Validator , GitHub - bids-standard/bids-validator: Validator for the Brain Imaging Data Structure ).
Best,
itreves
September 9, 2024, 2:51pm
8
I’m passing BIDS-validation, but I think there is something weird going on with the series where it is reading the AP in as fieldmaps. Here is my command (telling it to ignore fieldmaps):
$'Command :\n'singularity run \
-e \
-B /om2/scratch/tmp/treves/jhana_fmriprep/sub-002,/om2/user/treves/.cache \
/om2/user/treves/software/qsiprep-0.22.0.sif \
/om2/scratch/tmp/treves/jhana_fmriprep/sub-002/data \
/om2/scratch/tmp/treves/jhana_fmriprep/sub-002/data/derivatives_qsi/ \
participant \
--participant_label sub-002 \
-w /om2/scratch/tmp/treves/jhana_fmriprep/sub-002 \
--fs-license-file /om2/scratch/tmp/treves/jhana_fmriprep/sub-002/data/code/license.txt \
--unringing_method mrdegibbs \
--denoise-method patch2self \
--ignore fieldmaps \
--output_resolution 1.25 \
--skip-odf-reports \
--stop-on-first-crash \
--notrack \
--mem-mb 31500 \
--nthreads 16 \
--omp-nthreads 8 \
--recon-spec mrtrix_multishell_msmt_ACT-hsvs
Here is the offending (to my eyes) log info :
240909-10:38:54,895 nipype.workflow INFO:
Combining all 4 dwis within the single available session
240909-10:38:55,124 nipype.workflow INFO:
[{'dwi_series': ['/om2/scratch/tmp/treves/jhana_fmriprep/sub-002/data/sub-002/dwi/sub-002_acq-79_dir-PA_dwi.nii.gz', '/om2/scratch/tmp/treves/jhana_fmriprep/sub-002/data/sub-002/dwi/sub-002_acq-80_dir-PA_dwi.nii.gz'], 'dwi_series_pedir': 'j', 'fieldmap_info': {'suffix': 'rpe_series', 'rpe_series': ['/om2/scratch/tmp/treves/jhana_fmriprep/sub-002/data/sub-002/dwi/sub-002_acq-79_dir-AP_dwi.nii.gz', '/om2/scratch/tmp/treves/jhana_fmriprep/sub-002/data/sub-002/dwi/sub-002_acq-80_dir-AP_dwi.nii.gz']}, 'concatenated_bids_name': 'sub-002'}]
Here is the error:
UnboundLocalError: local variable 'dwi_denoise_window' referenced before assignment
Steven
September 9, 2024, 3:00pm
9
Hi @itreves ,
I see issues with your command.
It looks like you are inputting. a subject folder as input, whereas QSIPrep expects the BIDS root directory to be the input argument. I would reconsider how you are organizing data to one that makes more intuitive sense.
Not recommended to put working dir in BIDS dir
itreves:
--skip-odf-reports \
If you include --writable-tmpfs to the Apptainer preamble you can get the odf plots to work properly.
You do the HSVS workflow but do not specify a freesurfer input directory.
itreves:
Here is the offending (to my eyes) log info :
240909-10:38:54,895 nipype.workflow INFO:
Combining all 4 dwis within the single available session
240909-10:38:55,124 nipype.workflow INFO:
[{'dwi_series': ['/om2/scratch/tmp/treves/jhana_fmriprep/sub-002/data/sub-002/dwi/sub-002_acq-79_dir-PA_dwi.nii.gz', '/om2/scratch/tmp/treves/jhana_fmriprep/sub-002/data/sub-002/dwi/sub-002_acq-80_dir-PA_dwi.nii.gz'], 'dwi_series_pedir': 'j', 'fieldmap_info': {'suffix': 'rpe_series', 'rpe_series': ['/om2/scratch/tmp/treves/jhana_fmriprep/sub-002/data/sub-002/dwi/sub-002_acq-79_dir-AP_dwi.nii.gz', '/om2/scratch/tmp/treves/jhana_fmriprep/sub-002/data/sub-002/dwi/sub-002_acq-80_dir-AP_dwi.nii.gz']}, 'concatenated_bids_name': 'sub-002'}]
Nothing wrong there.
Can you also try upgrading to 0.22.1 or 0.23.0 just to make sure you are including all bug fixes? Of note, as of 0.23.0, qsiprep and qsirecon are split, so the recon-spec would be included in a separate qsirecon command. Perhaps also just try the default --denoise-method.
Best,
itreves
September 10, 2024, 2:14pm
10
I realize that my working directories are a little strange - I’m just running this on one subject so I think it’s okay for now. I revised my commands and got qsiprep to run, but not qsirecon.
$'Command :\n'singularity run \
--writable-tmpfs \
-e \
-B /om2/scratch/tmp/treves/jhana_fmriprep/sub-002,/om2/user/treves/.cache \
/om2/user/treves/software/qsiprep-0.22.1.sif \
/om2/scratch/tmp/treves/jhana_fmriprep/sub-002/data \
/om2/scratch/tmp/treves/jhana_fmriprep/sub-002/data/derivatives_qsi/ \
participant \
--participant_label sub-002 \
-w /om2/scratch/tmp/treves/jhana_fmriprep/sub-002 \
--fs-license-file /om2/scratch/tmp/treves/jhana_fmriprep/sub-002/data/code/license.txt \
--unringing_method mrdegibbs \
--ignore fieldmaps \
--output_resolution 1.25 \
--stop-on-first-crash \
--notrack \
--mem-mb 31500 \
--nthreads 16 \
--omp-nthreads 8 \
--recon-spec mrtrix_multishell_msmt_ACT-hsvs \
--skip_bids_validation \
--freesurfer-input /nese/mit/group/gablab/data/JhanaGroup/derivatives2/sub-002 \
--recon-only \
--recon-input /om2/scratch/tmp/treves/jhana_fmriprep/sub-002/data/derivatives_qsi/qsiprep
Can you check this? Is it an issue with the freesurfer processed data?log.txt (5.0 KB)
Steven
September 10, 2024, 2:35pm
11
Hi @itreves ,
It doesn’t look like you mounted the Freesurfer input in the Apptainer preamble.
Best,
itreves
September 10, 2024, 2:45pm
12
Hm, that didn’t fix it. Same error!
$'Command :\n'singularity run --writable-tmpfs -e -B /om2/scratch/tmp/treves/jhana_fmriprep/sub-002,/om2/user/treves/.cache,/nese/mit/group/gablab/data/JhanaGroup/derivatives2/sub-002 /om2/user/treves/software/qsiprep-0.22.1.sif /om2/scratch/tmp/treves/jhana_fmriprep/sub-002/data /om2/scratch/tmp/treves/jhana_fmriprep/sub-002/data/derivatives_qsi/ participant --participant_label sub-002 -w /om2/scratch/tmp/treves/jhana_fmriprep/sub-002 --fs-license-file /om2/scratch/tmp/treves/jhana_fmriprep/sub-002/data/code/license.txt --unringing_method mrdegibbs --ignore fieldmaps --output_resolution 1.25 --stop-on-first-crash --notrack --mem-mb 31500 --nthreads 16 --omp-nthreads 8 --recon-spec mrtrix_multishell_msmt_ACT-hsvs --skip_bids_validation --freesurfer-input /nese/mit/group/gablab/data/JhanaGroup/derivatives2/sub-002 --recon-only --recon-input /om2/scratch/tmp/treves/jhana_fmriprep/sub-002/data/derivatives_qsi/qsiprep
Steven
September 10, 2024, 2:48pm
13
Can you make the freesurfer input one directory above? So ending with derivatives2?
itreves
September 10, 2024, 8:49pm
14
That seems to be working, thanks! ‘tractography’ is taking a long time (~ 2 hours), is that to be expected? (7T scanner)
Steven
September 10, 2024, 8:54pm
15
Hi @itreves ,
Yes, as you can see here (qsirecon/qsirecon/data/pipelines/mrtrix_multishell_msmt_ACT-hsvs.json at 9f4d5feb92616790045527004785c5f33e2cfd50 · PennLINC/qsirecon · GitHub ) the tractography algorithm creates 10m streamlines, which can take a while. The magnet strength does not impact that time.
Best,