Dear All,
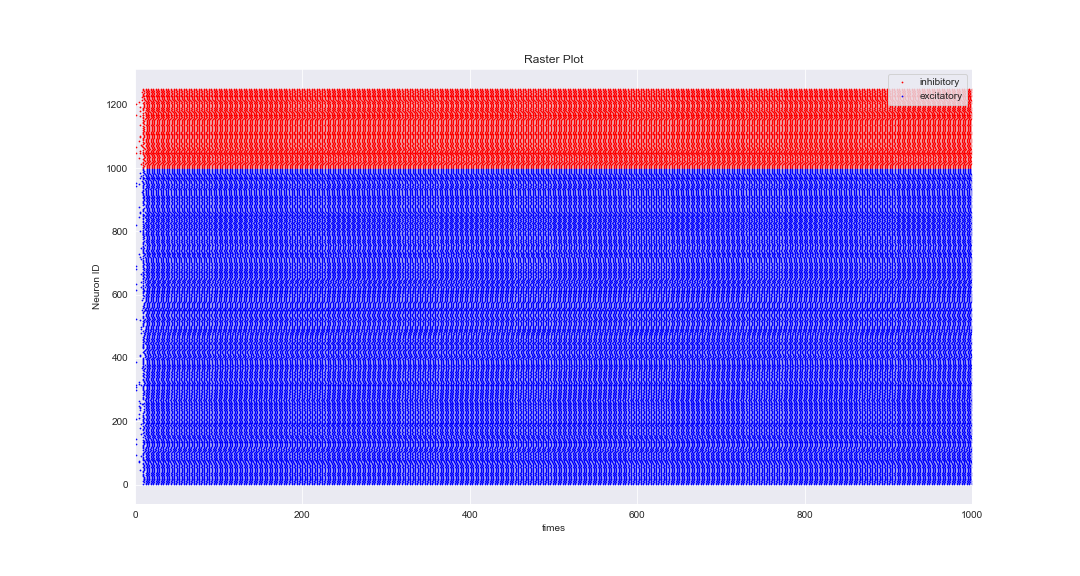
I tried to simulate an EI network using almost the same parameters as provided in @neuromatchacademy (W3D1) in the tutorial part but unfortunately, the network doesn’t generate natural rhythms. Here are all the parameters I used via NEST simulator:
"NE”, ”1000” #number of E neurons
"NI”, ”250” #number of I neurons
"CI”, ”25” #number of I connections
"CE”, ”100” #number of E connections
"tauMem”,”10.0” #membrane time constant
"J_ex”, 6 #excitatory connection weight
"J_in”, -36 #inhibitory connection weight
"p_rate”, 10 #poisson rate
"delay”, 2.5 #synaptic delay
"tau_syn_ex”,2 #synaptic decay time of E neurons
"tau_syn_in”,5 #synaptic decay time of I neurons
"V_th”, -55
“V_reset”,-75
"g_L”, 10
"V_init”, -65
"E_e”, 0
"E_I”, -80
"E_L”, -75
"tref”, 2
And this is the raster plot of 1000 ms of simulation:
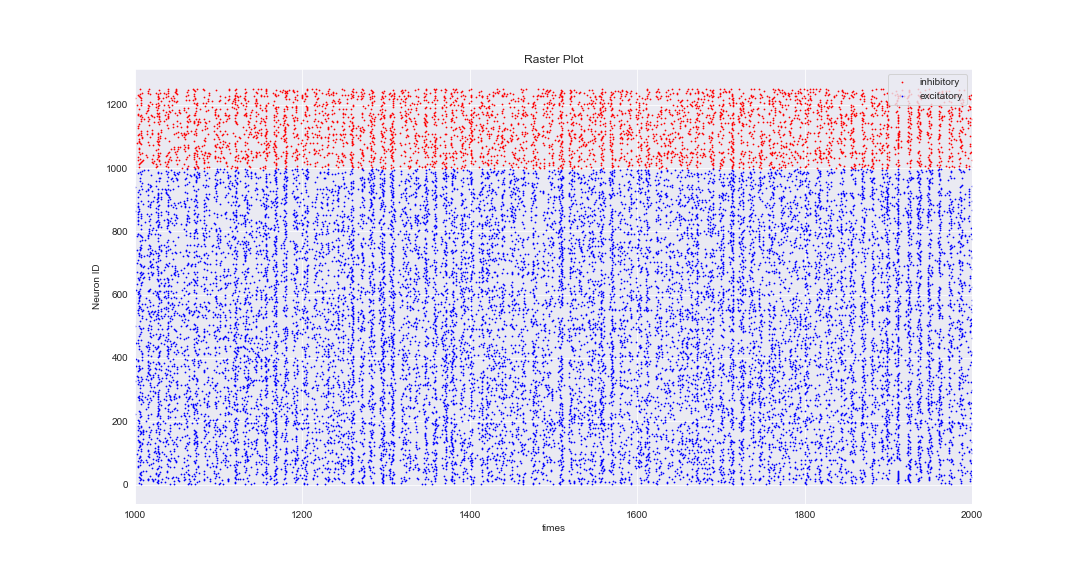
Notes: I tried to play with the parameters but I still get the same results. One of the ways that I could get realistic results was setting:
"tau_syn_ex”, 0.3 #synaptic decay time of E neurons
"tau_syn_in”, 0.5 #synaptic decay time of I neurons
But these parameters are not even close to biological parameters of neuron so I cannot use them really.
Can anyone tell me what’s that I’m doing it wrong, please?
Bests,
Nosratullah Mohammadi