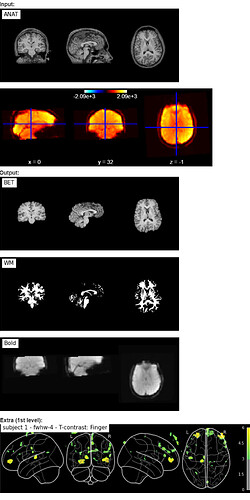
What happened?
Hello everyone,
I’ve tried to preprocess only one subject, but the procedure always fails. Please, see the output below.
I don’t know how to fix it.
Is my data BIDS valid?
Yes
What pulse sequence did I use?
Equipament: HDxt (1.5 T)
Anatomic (FSPGR)
TE: 3.124
TR: 8.248
Acquisition Matrix: 256x256
Bold
TE: 50
TR: 2500
Acquisition Matrix: 64x64
Relevant code:
extract = Node(ExtractROI(t_min=4, t_size=-1, output_type='NIFTI'),
name="extract")
mcflirt = Node(MCFLIRT(mean_vol=True,
save_plots=True,
output_type='NIFTI'),
name="mcflirt")
slicetimer = Node(SliceTimer(index_dir=False,
interleaved=True,
output_type='NIFTI',
time_repetition=TR),
name="slicetimer")
smooth = Node(Smooth(), name="smooth")
smooth.iterables = ("fwhm", fwhm)
art = Node(ArtifactDetect(norm_threshold=2,
zintensity_threshold=3,
mask_type='spm_global',
parameter_source='FSL',
use_differences=[True, False],
plot_type='svg'),
name="art")
[...]
bet_anat = Node(BET(frac=0.6,
robust=True,
output_type='NIFTI_GZ'),
name="bet_anat")
segmentation = Node(FAST(img_type = 1, bias_lowpass = 30,segment_iters = 50 , output_type='NIFTI_GZ'),
name="segmentation", mem_gb=4)
def get_wm(files):
return files[-1]
threshold = Node(Threshold(thresh=0.5,
args='-bin',
output_type='NIFTI_GZ'),
name="threshold")
coreg_pre = Node(FLIRT(dof=6, output_type='NIFTI_GZ'),
name="coreg_pre")
coreg_bbr = Node(FLIRT(dof=6,
cost='bbr',
schedule=opj(os.getenv('FSLDIR'),
'etc/flirtsch/bbr.sch'),
output_type='NIFTI_GZ'),
name="coreg_bbr")
applywarp = Node(FLIRT(interp='spline',
apply_isoxfm=iso_size,
output_type='NIFTI'),
name="applywarp")
applywarp_mean = Node(FLIRT(interp='spline',
apply_isoxfm=iso_size,
output_type='NIFTI_GZ'),
name="applywarp_mean")
coregwf = Workflow(name='coregwf')
coregwf.base_dir = opj(experiment_dir, working_dir)
coregwf.connect([(bet_anat, segmentation, [('out_file', 'in_files')]),
(segmentation, threshold, [(('partial_volume_files', get_wm),
'in_file')]),
(bet_anat, coreg_pre, [('out_file', 'reference')]),
(threshold, coreg_bbr, [('out_file', 'wm_seg')]),
(coreg_pre, coreg_bbr, [('out_matrix_file', 'in_matrix_file')]),
(coreg_bbr, applywarp, [('out_matrix_file', 'in_matrix_file')]),
(bet_anat, applywarp, [('out_file', 'reference')]),
(coreg_bbr, applywarp_mean, [('out_matrix_file', 'in_matrix_file')]),
(bet_anat, applywarp_mean, [('out_file', 'reference')]),
])
[...]
preproc = Workflow(name='preproc')
preproc.base_dir = opj(experiment_dir, working_dir)
preproc.connect([(infosource, selectfiles, [('subject_id', 'subject_id'),
('task_name', 'task_name')]),
(selectfiles, extract, [('func', 'in_file')]),
(extract, mcflirt, [('roi_file', 'in_file')]),
(mcflirt, slicetimer, [('out_file', 'in_file')]),
(selectfiles, coregwf, [('anat', 'bet_anat.in_file'),
('anat', 'coreg_bbr.reference')]),
(mcflirt, coregwf, [('mean_img', 'coreg_pre.in_file'),
('mean_img', 'coreg_bbr.in_file'),
('mean_img', 'applywarp_mean.in_file')]),
(slicetimer, coregwf, [('slice_time_corrected_file', 'applywarp.in_file')]),
(coregwf, smooth, [('applywarp.out_file', 'in_files')]),
(mcflirt, datasink, [('par_file', 'preproc.@par')]),
(smooth, datasink, [('smoothed_files', 'preproc.@smooth')]),
(coregwf, datasink, [('applywarp_mean.out_file', 'preproc.@mean')]),
(coregwf, art, [('applywarp.out_file', 'realigned_files')]),
(mcflirt, art, [('par_file', 'realignment_parameters')]),
(coregwf, datasink, [('coreg_bbr.out_matrix_file', 'preproc.@mat_file'),
('bet_anat.out_file', 'preproc.@brain')]),
(art, datasink, [('outlier_files', 'preproc.@outlier_files'),
('plot_files', 'preproc.@plot_files')]),
])