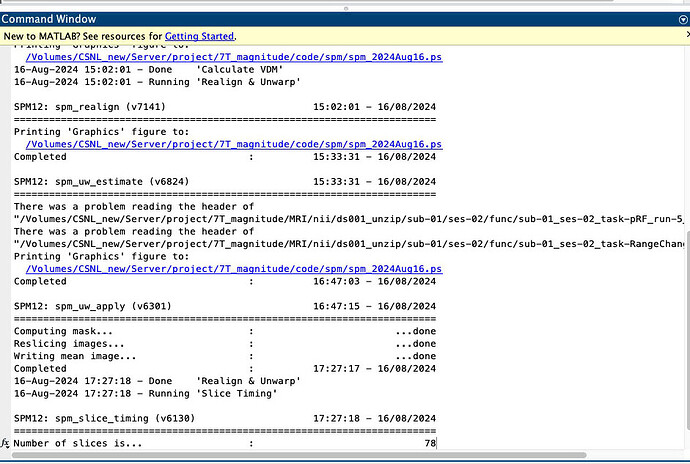
Hello, I am trying to proprocessing fMRI data by using spm12. I have been encountering with an error message “There was a problem reading the header of XXXX” like below.
Even though the messages appear at the command window, spm does not stop and proceeds. Therefore, I wonder what specific problem is involved in the error message, and can I ignore them. My batch code is like follows. I will appreciate your help!
nses = 3;
subs = 1:15;
for isub = subs
for ises = 1:nses
if isub < 10
ssub = sprintf('0%d',isub);
else
ssub = sprintf('%d',isub);
end
sses = sprintf('0%d',ises);
basedir0 = ['/Volumes/CSNL_new/Server/project/7T_magnitude/MRI/nii/ds001/sub-' ssub '/ses-' sses];
filename = {'anat','fmap','func'};
basedir = ['/Volumes/CSNL_new/Server/project/7T_magnitude/MRI/nii/ds001_unzip/sub-' ssub '/ses-' sses];
if isempty(dir(basedir))
mkdir([basedir '/' filename{1}])
mkdir([basedir '/' filename{2}])
mkdir([basedir '/' filename{3}])
end
subsesdir = ['sub-' ssub '_ses-' sses];
if isempty(dir([basedir '/anat/' subsesdir '_complete.mat'])) && isempty(dir([basedir '/anat/' subsesdir '_compute.mat']))
save([basedir '/anat/' subsesdir '_compute.mat'],'isub')
fprintf(['\n\n\n' subsesdir '\n\n\n'])
clear matlabbatch
% fieldmap correction
matlabbatch{1}.spm.tools.fieldmap.calculatevdm.subj.data.phasemag.shortphase = {[basedir '/fmap/' subsesdir '_phase1.nii,1']};
matlabbatch{1}.spm.tools.fieldmap.calculatevdm.subj.data.phasemag.shortmag = {[basedir '/fmap/' subsesdir '_magnitude1.nii,1']};
matlabbatch{1}.spm.tools.fieldmap.calculatevdm.subj.data.phasemag.longphase = {[basedir '/fmap/' subsesdir '_phase2.nii,1']};
matlabbatch{1}.spm.tools.fieldmap.calculatevdm.subj.data.phasemag.longmag = {[basedir '/fmap/' subsesdir '_magnitude2.nii,1']};
matlabbatch{1}.spm.tools.fieldmap.calculatevdm.subj.defaults.defaultsval.et = [3 6];
matlabbatch{1}.spm.tools.fieldmap.calculatevdm.subj.defaults.defaultsval.maskbrain = 1;
matlabbatch{1}.spm.tools.fieldmap.calculatevdm.subj.defaults.defaultsval.blipdir = -1;
matlabbatch{1}.spm.tools.fieldmap.calculatevdm.subj.defaults.defaultsval.tert = 43.68;
matlabbatch{1}.spm.tools.fieldmap.calculatevdm.subj.defaults.defaultsval.epifm = 0;
matlabbatch{1}.spm.tools.fieldmap.calculatevdm.subj.defaults.defaultsval.ajm = 0;
matlabbatch{1}.spm.tools.fieldmap.calculatevdm.subj.defaults.defaultsval.uflags.method = 'Mark3D';
matlabbatch{1}.spm.tools.fieldmap.calculatevdm.subj.defaults.defaultsval.uflags.fwhm = 10;
matlabbatch{1}.spm.tools.fieldmap.calculatevdm.subj.defaults.defaultsval.uflags.pad = 0;
matlabbatch{1}.spm.tools.fieldmap.calculatevdm.subj.defaults.defaultsval.uflags.ws = 1;
matlabbatch{1}.spm.tools.fieldmap.calculatevdm.subj.defaults.defaultsval.mflags.template = {'/Volumes/Data_CSNL/people/LHS/codes/subject_common/spm12/toolbox/FieldMap/T1.nii'};
matlabbatch{1}.spm.tools.fieldmap.calculatevdm.subj.defaults.defaultsval.mflags.fwhm = 5;
matlabbatch{1}.spm.tools.fieldmap.calculatevdm.subj.defaults.defaultsval.mflags.nerode = 2;
matlabbatch{1}.spm.tools.fieldmap.calculatevdm.subj.defaults.defaultsval.mflags.ndilate = 4;
matlabbatch{1}.spm.tools.fieldmap.calculatevdm.subj.defaults.defaultsval.mflags.thresh = 0.5;
matlabbatch{1}.spm.tools.fieldmap.calculatevdm.subj.defaults.defaultsval.mflags.reg = 0.02;
matlabbatch{1}.spm.tools.fieldmap.calculatevdm.subj.session.epi = {[basedir '/func/' subsesdir '_task-pRF_run-1_bold.nii,1']};
matlabbatch{1}.spm.tools.fieldmap.calculatevdm.subj.matchvdm = 1;
matlabbatch{1}.spm.tools.fieldmap.calculatevdm.subj.sessname = 'session';
matlabbatch{1}.spm.tools.fieldmap.calculatevdm.subj.writeunwarped = 1;
matlabbatch{1}.spm.tools.fieldmap.calculatevdm.subj.anat = '';
matlabbatch{1}.spm.tools.fieldmap.calculatevdm.subj.matchanat = 1;
% realignment
ntask = 3;
tasknames = {'GranuChange','pRF','RangeChange'}; % the first frame of the granularity task is the reference for the motion correction because T1 is imaged just before performing the granularity task
nruns = NaN(1,ntask);
for itask = 1:ntask
iniifiles = dir([basedir '/func/sub*' tasknames{itask} '*nii']);
nruns(itask) = length(iniifiles);
end
nframes = [240 163 163];
totalframe = sum(nframes.*nruns);
iscans = cell(totalframe,1);
cframe = 1;
for itask = 1:ntask
for irun = 1:nruns(itask)
for iframe = 1:nframes(itask)
iscans{cframe} = sprintf([basedir '/func/' subsesdir '_task-' tasknames{itask} '_run-%d_bold.nii,%d'],irun,iframe);
cframe = cframe + 1;
end
end
end
matlabbatch{2}.spm.spatial.realignunwarp.data.scans = iscans;
matlabbatch{2}.spm.spatial.realignunwarp.data.pmscan(1) = cfg_dep('Calculate VDM: Voxel displacement map (Subj 1, Session 1)', substruct('.','val', '{}',{1}, '.','val', '{}',{1}, '.','val', '{}',{1}, '.','val', '{}',{1}), substruct('()',{1}, '.','vdmfile', '{}',{1}));
matlabbatch{2}.spm.spatial.realignunwarp.eoptions.quality = 0.9;
matlabbatch{2}.spm.spatial.realignunwarp.eoptions.sep = 4;
matlabbatch{2}.spm.spatial.realignunwarp.eoptions.fwhm = 5;
matlabbatch{2}.spm.spatial.realignunwarp.eoptions.rtm = 0;
matlabbatch{2}.spm.spatial.realignunwarp.eoptions.einterp = 2;
matlabbatch{2}.spm.spatial.realignunwarp.eoptions.ewrap = [0 0 0];
matlabbatch{2}.spm.spatial.realignunwarp.eoptions.weight = '';
matlabbatch{2}.spm.spatial.realignunwarp.uweoptions.basfcn = [12 12];
matlabbatch{2}.spm.spatial.realignunwarp.uweoptions.regorder = 1;
matlabbatch{2}.spm.spatial.realignunwarp.uweoptions.lambda = 100000;
matlabbatch{2}.spm.spatial.realignunwarp.uweoptions.jm = 0;
matlabbatch{2}.spm.spatial.realignunwarp.uweoptions.fot = [4 5];
matlabbatch{2}.spm.spatial.realignunwarp.uweoptions.sot = [];
matlabbatch{2}.spm.spatial.realignunwarp.uweoptions.uwfwhm = 4;
matlabbatch{2}.spm.spatial.realignunwarp.uweoptions.rem = 1;
matlabbatch{2}.spm.spatial.realignunwarp.uweoptions.noi = 5;
matlabbatch{2}.spm.spatial.realignunwarp.uweoptions.expround = 'Average';
matlabbatch{2}.spm.spatial.realignunwarp.uwroptions.uwwhich = [2 1];
matlabbatch{2}.spm.spatial.realignunwarp.uwroptions.rinterp = 4;
matlabbatch{2}.spm.spatial.realignunwarp.uwroptions.wrap = [0 0 0];
matlabbatch{2}.spm.spatial.realignunwarp.uwroptions.mask = 1;
matlabbatch{2}.spm.spatial.realignunwarp.uwroptions.prefix = 'u';
% slice timing correction
matlabbatch{3}.spm.temporal.st.scans{1}(1) = cfg_dep('Realign & Unwarp: Unwarped Images (Sess 1)', substruct('.','val', '{}',{2}, '.','val', '{}',{1}, '.','val', '{}',{1}), substruct('.','sess', '()',{1}, '.','uwrfiles'));
matlabbatch{3}.spm.temporal.st.nslices = 78;
matlabbatch{3}.spm.temporal.st.tr = 1.5;
matlabbatch{3}.spm.temporal.st.ta = 1.4808;
matlabbatch{3}.spm.temporal.st.so = 1000*[1.095 0 0.75 0.0575 0.8075 0.115 0.865 0.1725 0.9225 0.23 0.98 0.2875 1.0375 0.4025 1.1525 0.46 1.21 0.5175 1.2675 0.575 1.325 0.6325 1.3825 0.69 1.44 0.345 1.095 0 0.75 0.0575 0.8075 0.115 0.865 0.1725 0.9225 0.23 0.98 0.2875 1.0375 0.4025 1.1525 0.46 1.21 0.5175 1.2675 0.575 1.325 0.6325 1.3825 0.69 1.44 0.345 1.095 0 0.75 0.0575 0.8075 0.115 0.865 0.1725 0.9225 0.23 0.98 0.2875 1.0375 0.4025 1.1525 0.46 1.21 0.5175 1.2675 0.575 1.325 0.6325 1.3825 0.69 1.44 0.345];
matlabbatch{3}.spm.temporal.st.refslice = 750;
matlabbatch{3}.spm.temporal.st.prefix = 'a';
% coregstration bold to the first session T1
if ises > 1
jbasedir = ['/Volumes/CSNL_new/Server/project/7T_magnitude/MRI/nii/ds001_unzip/sub-' ssub '/ses-01'];
matlabbatch{4}.spm.spatial.coreg.estimate.ref(1) = {[jbasedir '/anat/sub-' ssub '_ses-01_T1w.nii,1']};
matlabbatch{4}.spm.spatial.coreg.estimate.source = {[basedir '/anat/' subsesdir '_T1w.nii,1']}; % Assumption: native T1 ans functional images are aligned. So, coregistration is conducted only to the session-alignment
matlabbatch{4}.spm.spatial.coreg.estimate.other = cfg_dep('Slice Timing: Slice Timing Corr. Images (Sess 1)', substruct('.','val', '{}',{3}, '.','val', '{}',{1}, '.','val', '{}',{1}), substruct('()',{1}, '.','files'));
matlabbatch{4}.spm.spatial.coreg.estimate.eoptions.cost_fun = 'nmi';
matlabbatch{4}.spm.spatial.coreg.estimate.eoptions.sep = [4 2];
matlabbatch{4}.spm.spatial.coreg.estimate.eoptions.tol = [0.02 0.02 0.02 0.001 0.001 0.001 0.01 0.01 0.01 0.001 0.001 0.001];
matlabbatch{4}.spm.spatial.coreg.estimate.eoptions.fwhm = [7 7];
end
spm_jobman('run', matlabbatch);
delete([basedir '/anat/' subsesdir '_compute.mat'])
save([basedir '/anat/' subsesdir '_complete.mat'],'isub')
end
end
end