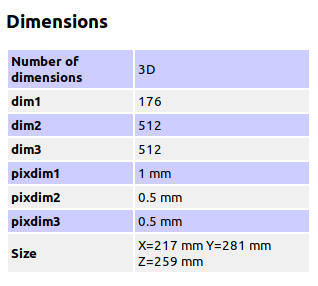
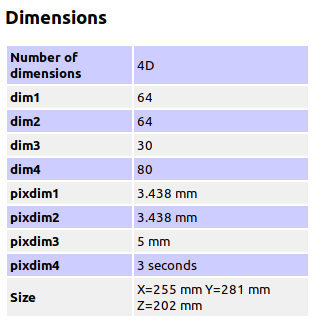
Summary of what happened:
I am encountering an error during the execution of the ‘syn’ node in FMRIPrep pipeline . The error message indicates an issue related to the file ‘fmap_syn0Warp.nii.gz’ in the specified path. I would like to understand better why this error is occurring during the execution of the ‘syn’ node. Is there anything specific about the settings or parameters that could be causing this problem? How can I diagnose and rectify this error in the context of SyN registration?
Command used (and if a helper script was used, a link to the helper script or the command generated):
#!/bin/bash
echo Start
for sub in 003 017 024 031 041 049 054 066 076 082 092 097 106 115 123 128; do
fmriprep-docker /media/pisa/0ABA/Elisa/bids /media/pisa/0ABA/Elisa/derivatives participant --participant-label ${sub} --fs-license-file /home/pisa/Desktop/license.txt --output-spaces MNI152NLin2009cAsym --stop-on-first-crash --write-graph --use-syn-sdc -w /media/pisa/0ABA/Elisa/workdir
done
Version:
fMRIPrep-23.1.4
Environment (Docker, Singularity, custom installation):
Docker
Data formatted according to a validatable standard? Please provide the output of the validator:
Yes
Relevant log outputs (up to 20 lines):
Node: fmriprep_23_1_wf.single_subject_002_wf.fmap_preproc_wf.wf_auto_00000.syn
Working directory: /scratch/fmriprep_23_1_wf/single_subject_002_wf/fmap_preproc_wf/wf_auto_00000/syn
Node inputs:
args = <undefined>
collapse_output_transforms = True
convergence_threshold = [1e-06, 1e-08]
convergence_window_size = [5, 2]
dimension = 3
environ = {'NSLOTS': '8'}
fixed_image = <undefined>
fixed_image_mask = <undefined>
fixed_image_masks = <undefined>
float = <undefined>
initial_moving_transform = <undefined>
initial_moving_transform_com = <undefined>
initialize_transforms_per_stage = False
interpolation = Linear
interpolation_parameters = <undefined>
invert_initial_moving_transform = <undefined>
metric = [['Mattes', 'Mattes'], ['Mattes', 'Mattes']]
metric_item_trait = <undefined>
metric_stage_trait = <undefined>
metric_weight = [[0.5, 0.5], [0.5, 0.5]]
metric_weight_item_trait = 1.0
metric_weight_stage_trait = <undefined>
moving_image = <undefined>
moving_image_mask = <undefined>
moving_image_masks = <undefined>
num_threads = 8
number_of_iterations = [[200, 100], [10]]
output_inverse_warped_image = False
output_transform_prefix = fmap_syn
output_warped_image = False
radius_bins_item_trait = 5
radius_bins_stage_trait = <undefined>
radius_or_number_of_bins = [[48, 48], [48, 48]]
random_seed = <undefined>
restore_state = <undefined>
restrict_deformation = <undefined>
sampling_percentage = [[0.8, 0.8], [1.0, 1.0]]
sampling_percentage_item_trait = <undefined>
sampling_percentage_stage_trait = <undefined>
sampling_strategy = [['Random', 'Random'], [None, None]]
sampling_strategy_item_trait = <undefined>
sampling_strategy_stage_trait = <undefined>
save_state = <undefined>
shrink_factors = [[1, 1], [1]]
sigma_units = ['vox', 'vox']
smoothing_sigmas = [[2.0, 0.0], [0.0]]
transform_parameters = [(0.8, 6.0, 3.0), (0.8, 2.0, 1.0)]
transforms = ['SyN', 'SyN']
use_estimate_learning_rate_once = <undefined>
use_histogram_matching = [True, True]
verbose = False
winsorize_lower_quantile = 0.001
winsorize_upper_quantile = 0.998
write_composite_transform = False
Traceback (most recent call last):
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/nipype/pipeline/plugins/multiproc.py", line 67, in run_node
result["result"] = node.run(updatehash=updatehash)
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py", line 527, in run
result = self._run_interface(execute=True)
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py", line 645, in _run_interface
return self._run_command(execute)
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py", line 771, in _run_command
raise NodeExecutionError(msg)
nipype.pipeline.engine.nodes.NodeExecutionError: Exception raised while executing Node syn.
Cmdline:
antsRegistration --collapse-output-transforms 1 --dimensionality 3 --initialize-transforms-per-stage 0 --interpolation Linear --output fmap_syn --transform SyN[ 0.8, 6.0, 3.0 ] --metric Mattes[ /scratch/fmriprep_23_1_wf/single_subject_002_wf/syn_preprocessing_auto_00000/anat2epi/clipped_noise_corrected_trans.nii.gz, /scratch/fmriprep_23_1_wf/single_subject_002_wf/fmap_preproc_wf/wf_auto_00000/clip_epi/clipped.nii.gz, 0.5, 48, Random, 0.8 ] --metric Mattes[ /scratch/fmriprep_23_1_wf/single_subject_002_wf/fmap_preproc_wf/wf_auto_00000/lap_anat_norm/clipped_noise_corrected_trans_maths_norm.nii.gz, /scratch/fmriprep_23_1_wf/single_subject_002_wf/fmap_preproc_wf/wf_auto_00000/lap_epi_norm/clipped_maths_norm.nii.gz, 0.5, 48, Random, 0.8 ] --convergence [ 200x100, 1e-06, 5 ] --smoothing-sigmas 2.0x0.0vox --shrink-factors 1x1 --use-histogram-matching 1 --restrict-deformation 0.1x1.0x0.1 --masks [ /scratch/fmriprep_23_1_wf/single_subject_002_wf/syn_preprocessing_auto_00000/mask2epi/sub-002_ses-01_T1w_noise_corrected_template_corrected_xform_rbrainmask_trans_uint8.nii, /scratch/fmriprep_23_1_wf/single_subject_002_wf/fmap_preproc_wf/wf_auto_00000/epi_umask/clipped_mask_union.nii.gz ] --transform SyN[ 0.8, 2.0, 1.0 ] --metric Mattes[ /scratch/fmriprep_23_1_wf/single_subject_002_wf/syn_preprocessing_auto_00000/anat2epi/clipped_noise_corrected_trans.nii.gz, /scratch/fmriprep_23_1_wf/single_subject_002_wf/fmap_preproc_wf/wf_auto_00000/clip_epi/clipped.nii.gz, 0.5, 48 ] --metric Mattes[ /scratch/fmriprep_23_1_wf/single_subject_002_wf/fmap_preproc_wf/wf_auto_00000/lap_anat_norm/clipped_noise_corrected_trans_maths_norm.nii.gz, /scratch/fmriprep_23_1_wf/single_subject_002_wf/fmap_preproc_wf/wf_auto_00000/lap_epi_norm/clipped_maths_norm.nii.gz, 0.5, 48 ] --convergence [ 10, 1e-08, 2 ] --smoothing-sigmas 0.0vox --shrink-factors 1 --use-histogram-matching 1 --restrict-deformation 0.1x1.0x0.1 --masks [ /scratch/fmriprep_23_1_wf/single_subject_002_wf/syn_preprocessing_auto_00000/mask2epi/sub-002_ses-01_T1w_noise_corrected_template_corrected_xform_rbrainmask_trans_uint8.nii, /scratch/fmriprep_23_1_wf/single_subject_002_wf/fmap_preproc_wf/wf_auto_00000/epi_umask/clipped_mask_union.nii.gz ] --winsorize-image-intensities [ 0.001, 0.998 ] --write-composite-transform 0
Stdout:
Stderr:
Killed
Traceback:
Traceback (most recent call last):
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/nipype/interfaces/base/core.py", line 453, in aggregate_outputs
setattr(outputs, key, val)
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/traits/trait_types.py", line 2699, in validate
return TraitListObject(self, object, name, value)
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/traits/trait_list_object.py", line 582, in __init__
super().__init__(
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/traits/trait_list_object.py", line 213, in __init__
super().__init__(self.item_validator(item) for item in iterable)
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/traits/trait_list_object.py", line 213, in <genexpr>
super().__init__(self.item_validator(item) for item in iterable)
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/traits/trait_list_object.py", line 865, in _item_validator
return trait_validator(object, self.name, value)
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/nipype/interfaces/base/traits_extension.py", line 330, in validate
value = super(File, self).validate(objekt, name, value, return_pathlike=True)
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/nipype/interfaces/base/traits_extension.py", line 135, in validate
self.error(objekt, name, str(value))
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/traits/base_trait_handler.py", line 74, in error
raise TraitError(
traits.trait_errors.TraitError: Each element of the 'reverse_forward_transforms' trait of a RegistrationOutputSpec instance must be a pathlike object or string representing an existing file, but a value of '/scratch/fmriprep_23_1_wf/single_subject_002_wf/fmap_preproc_wf/wf_auto_00000/syn/fmap_syn0Warp.nii.gz' <class 'str'> was specified.
During handling of the above exception, another exception occurred:
Traceback (most recent call last):
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/nipype/interfaces/base/core.py", line 400, in run
outputs = self.aggregate_outputs(runtime)
File "/opt/conda/envs/fmriprep/lib/python3.10/site-packages/nipype/interfaces/base/core.py", line 460, in aggregate_outputs
raise FileNotFoundError(msg)
FileNotFoundError: No such file or directory '['/scratch/fmriprep_23_1_wf/single_subject_002_wf/fmap_preproc_wf/wf_auto_00000/syn/fmap_syn0Warp.nii.gz']' for output 'reverse_forward_transforms' of a FixHeaderRegistration interface
Screenshots / relevant information:
├── dataset_description.json
├── sub-002
│ ├── ses-01
│ │ ├── anat
│ │ │ ├── sub-002_ses-01_T1w.json
│ │ │ └── sub-002_ses-01_T1w.nii
│ │ ├── dwi
│ │ │ ├── sub-002_ses-01_dwi.bval
│ │ │ ├── sub-002_ses-01_dwi.bvec
│ │ │ ├── sub-002_ses-01_dwi.json
│ │ │ └── sub-002_ses-01_dwi.nii
│ │ └── func
│ │ ├── sub-002_ses-01_task-rest_bold.json
│ │ └── sub-002_ses-01_task-rest_bold.nii
│ └── ses-02
│ ├── anat
│ │ ├── sub-002_ses-02_T1w.json
│ │ └── sub-002_ses-02_T1w.nii
│ ├── dwi
│ │ ├── sub-002_ses-02_dwi.bval
│ │ ├── sub-002_ses-02_dwi.bvec
│ │ ├── sub-002_ses-02_dwi.json
│ │ └── sub-002_ses-02_dwi.nii
│ └── func
│ ├── sub-002_ses-02_task-rest_bold.json
│ └── sub-002_ses-02_task-rest_bold.nii