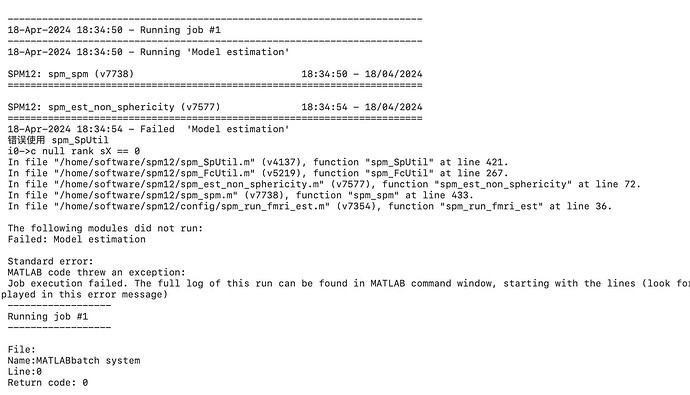
Summary of what happened:
Hi experts,
I am trying to do fMRI analysis with nipype on our lab HPC. Following the nipype tutorial, I did 1st level analysis successfully on my local M1 Mac with spm12, python3.10 and Matlab2020b. However, when it comes to HPC, spm function usage error occurred.
Command used (and if a helper script was used, a link to the helper script or the command generated):
from os.path import join as opj
import json
from nipype.interfaces.spm import Level1Design, EstimateModel, EstimateContrast
from nipype.algorithms.modelgen import SpecifySPMModel
from nipype.interfaces.utility import Function, IdentityInterface
from nipype.interfaces.io import SelectFiles, DataSink
from nipype import Workflow, Node, MapNode
# from nipype.interfaces.fsl import ExtractROI
from nipype.algorithms.misc import Gunzip
from nipype.interfaces.spm import Smooth
import argparse
# Specify which SPM to use
from nipype.interfaces import spm
spm.SPMCommand.set_mlab_paths(paths='/home/software/spm12')
# Get current user
import getpass
import os
user = getpass.getuser()
print('Running code as: ', user)
working_dir = '/home/{}/Guilt_Corr/fMRI/work/'.format(user)
data_dir = '/home/{}/Guilt_Corr/fMRI/derivatives/fmriprep/'.format(user) # BIDS main
exp_dir = '/home/{}/Guilt_Corr/fMRI/derivatives/first_level/'.format(user)
output_dir = os.path.join(exp_dir + "GBinteraction/") # Output for analyses
experiment_dir = data_dir
subject_list = ['sub-001']
TR = 1.0
# Build Workflow
# 1) Unzip functional image and brain mask
gunzip = MapNode(Gunzip(), name="gunzip", iterfield=['in_file'])
mask_gunzip = Node(Gunzip(), name="mask_gunzip", iterfield=['in_file'])
# 2) Drop dummy scans
# extract = MapNode(
# ExtractROI(t_min=0, t_size=-1, output_type="NIFTI"),
# iterfield=["in_file"],
# name="extractROI")
# 3) SpecifyModel - Generates SPM-specific Model
modelspec = Node(SpecifySPMModel(concatenate_runs=False,
input_units='secs',
output_units='secs',
time_repetition=TR,
high_pass_filter_cutoff=128),
name="modelspec")
# 4) Level1Design - Generates an SPM design matrix
level1design = Node(Level1Design(bases={'hrf': {'derivs': [1, 0]}},
timing_units='secs',
interscan_interval=TR,
model_serial_correlations='AR(1)'),
name="level1design")
# 5) EstimateModel - estimate the parameters of the model
level1estimate = Node(EstimateModel(estimation_method={'Classical': 1}),
name="level1estimate")
# 6) EstimateContrast - estimates contrasts
level1conest = Node(EstimateContrast(), name="level1conest")
# 7) Condition names & Contrasts
condition_names = ['HV','HN','LV','LN','Filler']
# Canonical Contrasts
cont01 = ['highGuilt_victimBriber', 'T', condition_names, [1, 0, 0, 0, 0]]
cont02 = ['highGuilt_nonvictimBriber', 'T', condition_names, [0, 1, 0, 0, 0]]
cont03 = ['lowGuilt_victimBriber','T', condition_names, [0, 0, 1, 0, 0]]
cont04 = ['lowGuilt_nonvictimBriber','T', condition_names, [0, 0, 0, 1, 0]]
cont05 = ['nonGuilt_Filler','T', condition_names, [0, 0, 0, 0, 1]]
# Main effects Contrasts
cont06 = ['Guilt_high > Guilt_low','T', condition_names, [1, 1, -1, -1, 0]]
cont07 = ['Briber_vict > Briber_nonvict','T', condition_names, [1, -1, 1, -1, 0]]
# Interaction effects Contrasts
cont08 = ['interaction','T', condition_names, [1, -1, -1, 1, 0]]
contrast_list = [cont01, cont02, cont03, cont04, cont05,
cont06, cont07,
cont08]
# 9) Function to get Subject specific condition information
def get_subject_info(subject_id):
from os.path import join as opj
import pandas as pd
onset_path = '/home/qiushiwei/Guilt_Corr/fMRI/mk_event_file/event_files/'
nr_path = '/home/qiushiwei/Guilt_Corr/fMRI/derivatives/fmriprep/%s'%subject_id
subjectinfo = []
if subject_id == "sub-042":
runs = ['1', '2']
else:
runs = ['1', '2', '3', '4']
for run in runs:
onset_file = opj(onset_path, '%s_task-guiltcorr_run-%s_events.tsv'%(subject_id, run))
ev = pd.read_csv(onset_file, sep="\t", usecols=['decision_onset','decision_duration','condition'])
regressor_file = opj(nr_path, 'func/%s_task-guiltcorr_run-%s_desc-confounds_timeseries.tsv'%(subject_id, run))
nuisance_reg = ['dvars', 'framewise_displacement'] + \
['a_comp_cor_%02d' % i for i in range(6)] + ['cosine%02d' % i for i in range(4)]
nr = pd.read_csv(regressor_file, sep="\t", usecols=nuisance_reg)
from nipype.interfaces.base import Bunch
run_info = Bunch(onsets=[], durations=[])
run_info.set(conditions=[g[0] for g in ev.groupby("condition")])
run_info.set(regressor_names=nr.columns.tolist())
run_info.set(regressors=nr.T.values.tolist())
for group in ev.groupby("condition"):
run_info.onsets.append(group[1].decision_onset.tolist())
run_info.durations.append(group[1].decision_duration.tolist())
subjectinfo.insert(int(run)-1,
run_info)
return subjectinfo
getsubjectinfo = Node(Function(input_names=['subject_id'],
output_names=['subject_info'],
function=get_subject_info),
name='getsubjectinfo')
# Infosource - a function free node to iterate over the list of subject names
infosource = Node(IdentityInterface(fields=['subject_id','contrasts'],
contrasts=contrast_list),
name="infosource")
infosource.iterables = [('subject_id', subject_list)]
# SelectFiles - to grab the data (alternativ to DataGrabber)
templates = {'func': '/home/qiushiwei/Guilt_Corr/fMRI/derivatives/fmriprep/{subject_id}/func/{subject_id}_task-guiltcorr_run-*_space-MNI152NLin2009cAsym_desc-preproc_bold.nii.gz',
'mask': '/home/qiushiwei/Guilt_Corr/fMRI/derivatives/fmriprep/{subject_id}/anat/{subject_id}_space-MNI152NLin2009cAsym_desc-brain_mask.nii.gz'}
selectfiles = Node(SelectFiles(templates,
base_directory=experiment_dir,
sort_filelist=True),
name="selectfiles")
# Datasink - creates output folder for important outputs
datasink = Node(DataSink(base_directory=experiment_dir,
container=output_dir),
name="datasink")
# Use the following DataSink output substitutions
substitutions = [('_subject_id_', ''),
('', '')]
datasink.inputs.substitutions = substitutions
subjFolders = [('sub-%s' % sub)
for sub in subject_list]
substitutions.extend(subjFolders)
# Initiation of the 1st-level analysis workflow
l1analysis = Workflow(name='1st_lv_workflow')
l1analysis.base_dir = opj(experiment_dir, working_dir)
# Connect up the 1st-level analysis components
l1analysis.connect([(infosource, selectfiles, [('subject_id', 'subject_id')]),
(infosource, getsubjectinfo, [('subject_id',
'subject_id')]),
(getsubjectinfo, modelspec, [('subject_info',
'subject_info')]),
(infosource, level1conest, [('contrasts', 'contrasts')]),
(selectfiles, gunzip, [('func', 'in_file')]),
(selectfiles, mask_gunzip, [('mask', 'in_file')]),
(gunzip, modelspec, [('out_file', 'functional_runs')]),
(modelspec, level1design, [('session_info',
'session_info')]),
(mask_gunzip, level1design, [('out_file',
'mask_image')]),
(level1design, level1estimate, [('spm_mat_file',
'spm_mat_file')]),
(level1estimate, level1conest, [('spm_mat_file',
'spm_mat_file'),
('beta_images',
'beta_images'),
('residual_image',
'residual_image')]),
(level1conest, datasink, [('spm_mat_file', '1st_lv_unsmoothed_avgcond.@spm_mat'),
('spmT_images', '1st_lv_unsmoothed_avgcond.@T'),
('con_images', '1st_lv_unsmoothed_avgcond.@con'),
('spmF_images', '1st_lv_unsmoothed_avgcond.@F'),
('ess_images', '1st_lv_unsmoothed_avgcond.@ess'),
]),
])
l1analysis.config["execution"]["crashfile_format"] = "txt"
l1analysis.run('MultiProc', plugin_args={'n_procs': 7})
Version:
HPC Linux Ubuntu 20.04
conda env python3.10
matlab 2022b
nipype 1.8.6
Environment (Docker, Singularity / Apptainer, custom installation):
conda installData formatted according to a validatable standard? Please provide the output of the validator:
BIDS
Relevant log outputs (up to 20 lines):
Traceback (most recent call last):
File "/home/qiushiwei/Guilt_Corr/fMRI/code/first_level_analysis_GB.py", line 218, in <module>
l1analysis.run('MultiProc', plugin_args={'n_procs': 7})
File "/home/qiushiwei/.conda/envs/nipype/lib/python3.10/site-packages/nipype/pipeline/engine/workflows.py", line 638, in run
runner.run(execgraph, updatehash=updatehash, config=self.config)
File "/home/qiushiwei/.conda/envs/nipype/lib/python3.10/site-packages/nipype/pipeline/plugins/base.py", line 224, in run
raise error from cause
RuntimeError: Traceback (most recent call last):
File "/home/qiushiwei/.conda/envs/nipype/lib/python3.10/site-packages/nipype/pipeline/plugins/multiproc.py", line 67, in run_node
result["result"] = node.run(updatehash=updatehash)
File "/home/qiushiwei/.conda/envs/nipype/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py", line 527, in run
result = self._run_interface(execute=True)
File "/home/qiushiwei/.conda/envs/nipype/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py", line 645, in _run_interface
return self._run_command(execute)
File "/home/qiushiwei/.conda/envs/nipype/lib/python3.10/site-packages/nipype/pipeline/engine/nodes.py", line 771, in _run_command
raise NodeExecutionError(msg)
nipype.pipeline.engine.nodes.NodeExecutionError: Exception raised while executing Node level1estimate.
Traceback:
Traceback (most recent call last):
File "/home/qiushiwei/.conda/envs/nipype/lib/python3.10/site-packages/nipype/interfaces/base/core.py", line 397, in run
runtime = self._run_interface(runtime)
File "/home/qiushiwei/.conda/envs/nipype/lib/python3.10/site-packages/nipype/interfaces/spm/base.py", line 386, in _run_interface
results = self.mlab.run()
File "/home/qiushiwei/.conda/envs/nipype/lib/python3.10/site-packages/nipype/interfaces/base/core.py", line 397, in run
runtime = self._run_interface(runtime)
File "/home/qiushiwei/.conda/envs/nipype/lib/python3.10/site-packages/nipype/interfaces/matlab.py", line 164, in _run_interface
self.raise_exception(runtime)
File "/home/qiushiwei/.conda/envs/nipype/lib/python3.10/site-packages/nipype/interfaces/base/core.py", line 685, in raise_exception
raise RuntimeError(
RuntimeError: Command:
matlab -nodesktop -nosplash -singleCompThread -r "addpath('/home/qiushiwei/Guilt_Corr/fMRI/work/1st_lv_workflow/_subject_id_sub-001/level1estimate');pyscript_estimatemodel;exit"
Standard output:
MATLAB is selecting SOFTWARE OPENGL rendering.