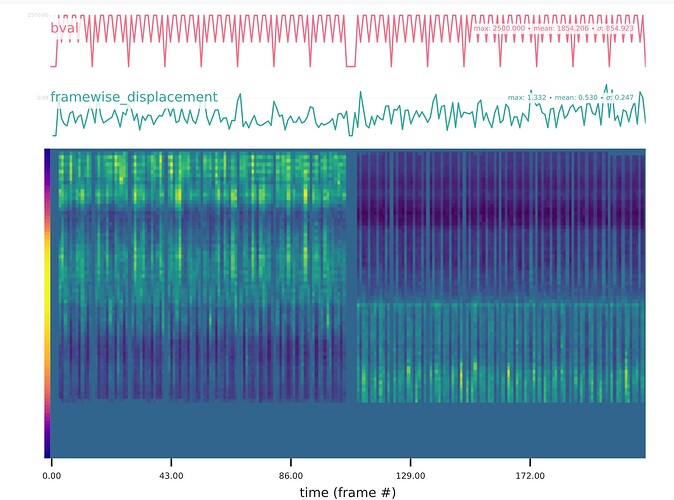
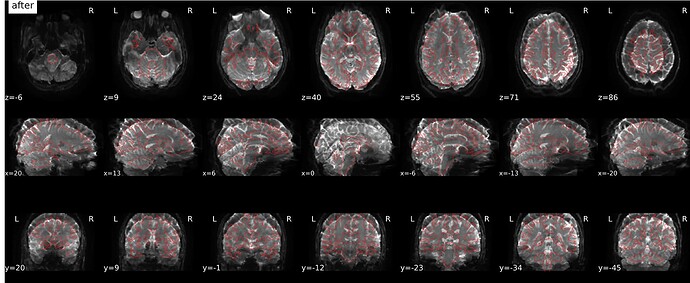
Summary of what happened:
We acquired a multi-shell diffusion sequence with two phase encoding directions (AP & PA). We tried running QSIPrep on a subject’s data, but encountered some errors which we unsure how to resolve. We would greatly appreciate any guidance from the QSIPrep community on how to proceed.
One error is related to the FOVs of the images. We see an open issue about this in QSIPrep’s GitHub Repo (see QSIPrep#273). To fix the error, we transferred the affine’s from the PA images to the AP images, as suggested here. Is this still the recommended approach, and are there any implications we should consider before proceeding? The comment also suggests using the --denoise-before-combining flag, but this doesn’t seem to be a valid QSIPrep flag.
Additionally, the documentation for HCP-style sequences recommends using both the --distortion-group-merge average and --combine-all-dwis flags. However, the second also does not appear to be valid.
Any clarification on the right approach to handle this FOV error and which flags to include would be much appreciated!
Command used (and if a helper script was used, a link to the helper script or the command generated):
qsiprep.sbatch
#!/bin/bash
#SBATCH -J qsiprep
#SBATCH --time=1-00:00:00
#SBATCH -n 1
#SBATCH --array 1 # TODO: update to include more subjects
#SBATCH --cpus-per-task=8
#SBATCH --mem-per-cpu=8G
#SBATCH -p russpold,hns,normal
# Outputs ----------------------------------
#SBATCH -o ./log/%x-%A-%a.out
#SBATCH -e ./log/%x-%A-%a.err
#SBATCH --mail-user=logben@stanford.edu
#SBATCH --mail-type=END
# ------------------------------------------
: '
Name: qsiprep.sbatch
Description: Submits a single batch QSIPrep job for each subject,
where the subject is determined by the line number in subs.txt
corresponding to the SLURM array ID.
'
# Prepare paths
source ./setup_env.sh
# Prepare apptainer/singularity command line
APPTAINER_CMD="apptainer run --cleanenv -B $BIDS_DIR:/data -B $WORK_DIR:/work $APPTAINER_IMAGE"
# Compose subject ID for each array task
SUBJECT=$( sed "${SLURM_ARRAY_TASK_ID}q;d" ${SUBJECTS_FILE} )
# Compose the command line with flags for QSIPrep
# NOTE: Include: --distortion-group-merge average?
cmd="$APPTAINER_CMD /data $DERIVS_DIR participant --participant-label $SUBJECT -w /work --output-resolution 1.5 --skip-bids-validation --verbose"
# Show and execute the command
echo Running task ${SLURM_ARRAY_TASK_ID}
echo Commandline: $cmd
eval $cmd
exitcode=$? # store exit code
echo Finished tasks ${SLURM_ARRAY_TASK_ID} with exit code $exitcode
exit $exitcode
setup_env.sh
#!/bin/bash
export VERSION="1.0.0rc1"
export IMAGE_NAME="qsiprep_${VERSION}"
export APPTAINER_IMAGE="${SCRATCH}/.apptainer/${IMAGE_NAME}.sif"
export PROJECT_DIR="${SCRATCH}/projects/rdoc_fmri"
export BIDS_DIR="${PROJECT_DIR}/data/bids"
export DERIVS_DIR="${BIDS_DIR}/derivatives/${IMAGE_NAME}"
export WORK_DIR="${PROJECT_DIR}/work/${IMAGE_NAME}"
export SUBJECTS_FILE="${PROJECT_DIR}/subs.txt"
mkdir -p "${DERIVS_DIR}"
mkdir -p "${WORK_DIR}"
Version:
QSIPrep version 1.0.0rc1 (docker pull pennlinc/qsiprep:1.0.0rc1)
Environment (Docker, Singularity / Apptainer, custom installation):
We are using apptainer and submitting the job on HPC.
Data formatted according to a validatable standard? Please provide the output of the validator:
Yes, the directory is in valid BIDS format. We received one warning about SliceTiming in one of our functional scans, which we chose to ignore:
bids-validator@1.14.6
1: [WARN] You should define 'SliceTiming' for this file. If you don't provide this information slice time correction will not be possible. 'Slice Timing' is the time at which each slice was acquired within each volume (frame) of the acquisition. Slice timing is not slice order -- rather, it is a list of times containing the time (in seconds) of each slice acquisition in relation to the beginning of volume acquisition. (code: 13 - SLICE_TIMING_NOT_DEFINED)
./sub-S1/ses-01/func/sub-S1_ses-01_task-rest_run-1_echo-1_bold.nii.gz
./sub-S1/ses-01/func/sub-S1_ses-01_task-rest_run-1_echo-2_bold.nii.gz
./sub-S1/ses-01/func/sub-S1_ses-01_task-rest_run-1_echo-3_bold.nii.gz
Relevant log outputs (up to 20 lines):
ValueError: Field of view of image #1 is different from reference FOV.
Reference affine:
array([[ -1.5 , 0. , -0. , 105.15640259],
[ -0. , 1.5 , -0. , -118.27559662],
[ 0. , 0. , 1.5 , -21.9727993 ],
[ 0. , 0. , 0. , 1. ]])
Image affine:
array([[ -1.5 , 0. , -0. , 104.05000305],
[ -0. , 1.5 , -0. , -105.34999847],
[ 0. , 0. , 1.5 , -43.59999847],
[ 0. , 0. , 0. , 1. ]])
Reference shape:
(140, 140, 84)
Image shape:
(140, 140, 84, 107)
Screenshots / relevant information:
This is the directory structure of our BIDS dataset:
├── dataset_description.json
├── fieldmap.json
├── README.md
└── sub-S1
├── anat
│ ├── sub-S1_run-1_T1w.json
│ ├── sub-S1_run-1_T1w.nii.gz
│ ├── sub-S1_run-1_T2w.json
│ └── sub-S1_run-1_T2w.nii.gz
├── dwi
│ ├── sub-S1_acq-G105_dir-AP_run-1_dwi.bval
│ ├── sub-S1_acq-G105_dir-AP_run-1_dwi.bvec
│ ├── sub-S1_acq-G105_dir-AP_run-1_dwi.json
│ ├── sub-S1_acq-G105_dir-AP_run-1_dwi.nii.gz
│ ├── sub-S1_acq-G105_dir-PA_run-1_dwi.bval
│ ├── sub-S1_acq-G105_dir-PA_run-1_dwi.bvec
│ ├── sub-S1_acq-G105_dir-PA_run-1_dwi.json
│ └── sub-S1_acq-G105_dir-PA_run-1_dwi.nii.gz
└── ses-01
├── fmap
│ ├── sub-S1_ses-01_run-1_fieldmap.json
│ ├── sub-S1_ses-01_run-1_fieldmap.nii.gz
│ └── sub-S1_ses-01_run-1_magnitude.nii.gz
└── func
├── sub-S1_ses-01_task-rest_run-1_echo-1_bold.json
├── sub-S1_ses-01_task-rest_run-1_echo-1_bold.nii.gz
├── sub-S1_ses-01_task-rest_run-1_echo-2_bold.json
├── sub-S1_ses-01_task-rest_run-1_echo-2_bold.nii.gz
├── sub-S1_ses-01_task-rest_run-1_echo-3_bold.json
└── sub-S1_ses-01_task-rest_run-1_echo-3_bold.nii.gz