Hello,
I am building a simple tool that helps remove respiration and cardiac noise from fMRI data based on RETROICOR noise model. I collected respiration signal and cardiac signal during the scanning, and already calculated the regressors values. And I have applied these regressors to FSL feat model with GUI which worked well. However, I couldn’t build contrast with Nipype. Hope if anyone has similar experience in building the contrasts of regressors could give me some advice. Big thanks!! ![]()
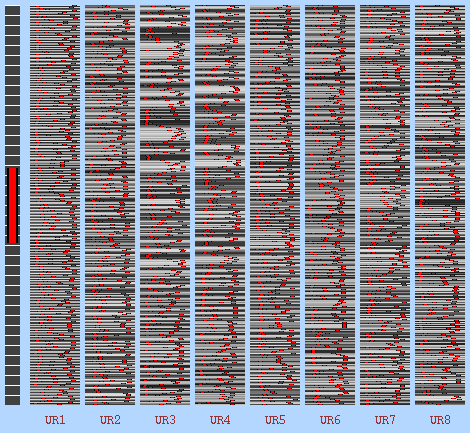
This is the contrast I have built with GUI of FSL, the first eight EVs are the calculated noise regressors and the rest is motion confounds. This worked well and give back t-stat results.
When I tried with Nipype, the FEAT model worked well without conrtasts, and it gave back parameter estimate files for each regressor. But when I tried to add regressors as contrasts, it gave back the warning that ‘the contrast is empty.’
The codes for specifying:
info = [Bunch(
conditions = [],
onsets = [],
durations = [],
regressors = [list(confounds['0']),
list(confounds['1']),
list(confounds['2']),
list(confounds['3']),
list(confounds['4']),
list(confounds['5']),
list(confounds['6']),
list(confounds['7']),
],
regressors_names = ['L0','L1','L2','L3','L4','L5','L6','L7']
)]
s = model.SpecifyModel()
s.inputs.input_units = 'secs'
s.inputs.time_repetition = 1.7
s.inputs.high_pass_filter_cutoff = 128
s.inputs.functional_runs = img
s.inputs.subject_info = info
specify_model_results = s.run()
Adding contrast:
reg_names = ['L0','L1','L2','L3','L4','L5','L6','L7']
cont_1 = ['ev0','T', reg_names,[1.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0, 0.0]]
contrasts = [cont_1]
level1design = mem.cache(fsl.model.Level1Design)
level1design_results = level1design(interscan_interval = 1.7,
bases = {'none':{'derivs': False}},
session_info = specify_model_results.outputs.session_info,
model_serial_correlations=False,
contrasts = contrasts
)
Failed in building the FEAT model. Codes with errors:
modelgen = mem.cache(fsl.model.FEATModel)
modelgen_results = modelgen(fsf_file=level1design_results.outputs.fsf_files,
ev_files=level1design_results.outputs.ev_files)
modelgen_results.outputs
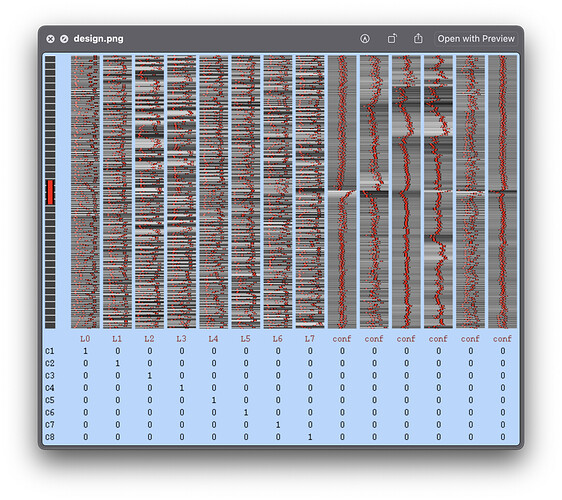
When I removed the contrast from FEAT, the design could be generated like this: