Hello all,
I am attempting to extract time series data through a parcellation using fslmeants
My residualized data is from a task that has been run through FSL feat and XCP engine (it’s some older data).
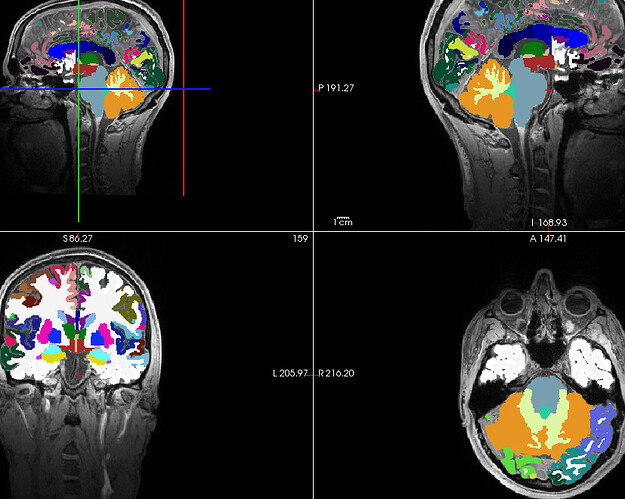
My parcellation looks like the following image (registered and oriented to the t1w)
My residualized data has the following specs:
type: nii
dimensions: 50 x 64 x 51 x 960
voxel sizes: 3.000000, 3.000000, 3.000000
type: FLOAT (3)
fov: 150.000
dof: 1
xstart: -75.0, xend: 75.0
ystart: -96.0, yend: 96.0
zstart: -76.5, zend: 76.5
TR: 0.00 msec, TE: 0.00 msec, TI: 0.00 msec, flip angle: 0.00 degrees
nframes: 960
PhEncDir: UNKNOWN
FieldStrength: 0.000000
ras xform present
xform info: x_r = 1.0000, y_r = 0.0000, z_r = 0.0000, c_r = 8.9143
: x_a = 0.0000, y_a = 1.0000, z_a = 0.0000, c_a = 5.2080
: x_s = 0.0000, y_s = 0.0000, z_s = 1.0000, c_s = -8.4251
Orientation : RAS
Primary Slice Direction: axial
voxel to ras transform:
3.0000 0.0000 0.0000 -66.0857
0.0000 3.0000 0.0000 -90.7920
0.0000 0.0000 3.0000 -84.9251
0.0000 0.0000 0.0000 1.0000
voxel-to-ras determinant 27
ras to voxel transform:
0.3333 0.0000 0.0000 22.0286
0.0000 0.3333 0.0000 30.2640
0.0000 0.0000 0.3333 28.3084
0.0000 0.0000 0.0000 1.0000
I then registered my parcellation to the res4d data using
antsRegistrationSyN.sh
-d 3 \
-n 25 \
-o ${reg_dir}/${sub}_seg2mask \
-f ${seg_oriented} \
-m ${mask} \
2>a1 > ${logfile}
antsApplyTransforms \
-i ${seg_oriented} \
-r ${mask} \
-o ${reg_dir}/${sub}_${task}_warped_seg2mask.nii.gz \
-n NearestNeighbor \
-t ${reg_dir}/${sub}_seg2maskWarped.nii.gz
And finally extract the mean time series from each roi in the segmentation:
fslmeants -i ${ts_img} \
-o ${ts_dir}/${sub}_${task}_${level}_li2019_ts.txt \
--label=${reg_dir}/${sub}_${task}_warped_seg2mask.nii.gz
However, the resulting time-series txt file is incorrect as it gives me the time-series for 1 region (potentially the whole brain) in a N x 1 matrix.
Any advice would be very much appreciated.